Abstract
The information about when and where each gene is to be expressed is mainly encoded in the DNA sequence of enhancers, sequence elements that comprise binding sites (motifs) for different transcription factors (TFs). Most of the research on enhancer sequences has been focused on TF motif presence, whereas the enhancer syntax, that is, the flexibility of important motif positions and how the sequence context modulates the activity of TF motifs, remains poorly understood. Here, we explore the rules of enhancer syntax by a two-pronged approach in Drosophila melanogaster S2 cells: we (1) replace important TF motifs by all possible 65,536 eight-nucleotide-long sequences and (2) paste eight important TF motif types into 763 positions within 496 enhancers. These complementary strategies reveal that enhancers display constrained sequence flexibility and the context-specific modulation of motif function. Important motifs can be functionally replaced by hundreds of sequences constituting several distinct motif types, but these are only a fraction of all possible sequences and motif types. Moreover, TF motifs contribute with different intrinsic strengths that are strongly modulated by the enhancer sequence context (the flanking sequence, the presence and diversity of other motif types, and the distance between motifs), such that not all motif types can work in all positions. The context-specific modulation of motif function is also a hallmark of human enhancers, as we demonstrate experimentally. Overall, these two general principles of enhancer sequences are important to understand and predict enhancer function during development, evolution, and in disease.
Transcriptional enhancers are DNA sequence elements that control gene expression by modulating the transcription of their target genes in specific cell types and conditions (Banerji et al. 1981; Levine 2010). These elements contain short sequence motifs bound by different transcription factors (TFs), and the combined regulatory cues of all bound TFs determine an enhancer's activity (Spitz and Furlong 2012). Due to the critical role of enhancers in development, evolution, and disease (Levine 2010; Rickels and Shilatifard 2018), understanding how enhancer sequences encode function is a major question in biology. Previous studies have highlighted the importance of sequence constraints within enhancers, such as the presence of TF motifs and features related to the motifs’ flanking sequences, affinities, and arrangements (their number, order, orientation, and spacing), termed here “motif syntax” (Jindal and Farley 2021). However, although mutations in enhancer sequences can change enhancer function and lead to morphological evolution and disease (Gompel et al. 2005; Visel et al. 2009; Levine 2010; Rickels and Shilatifard 2018), enhancers usually display only modest or no sequence conservation across species (Ludwig et al. 1998; Blow et al. 2010; Schmidt et al. 2010; May et al. 2012; Arnold et al. 2014; Villar et al. 2015; Fuqua et al. 2020) and even random DNA sequences can act as enhancers (de Boer et al. 2020; Galupa et al. 2023). Therefore, the importance of sequence constraints and motif syntax within enhancers remain outstanding questions in gene regulation.
Two main models have been proposed to explain how enhancer sequence relates to function. The enhanceosome model assumes very strict syntax rules with invariant motif arrangements required for cooperative TF binding (Thanos and Maniatis 1995; Panne 2008). In contrast, the billboard model proposes that TFs bind independently without constraints on how motifs are arranged within the enhancer (Kulkarni and Arnosti 2003; Arnosti and Kulkarni 2005). Yet very few enhancers fit these models, having either invariant syntax or no constraints at all, and most enhancers fall in between these two extremes, with a flexible syntax yet high degree of dependency between enhancer features (Kulkarni and Arnosti 2003; Vockley et al. 2017; Jindal and Farley 2021). This complexity in enhancer sequence has prevented the generalization of sequence rules derived from individual enhancers into unifying principles of the regulatory code, thus limiting our understanding of the sequence constraints related to motif syntax and TF activity in enhancers.
Although enhancer sequences evolve rapidly, their function, which is comprised of enhancer strength as well as cell type–specificity, can be conserved despite significant sequence changes (Ludwig et al. 1998, 2000; Rastegar et al. 2008; Blow et al. 2010; Schmidt et al. 2010; Weirauch and Hughes 2010; Swanson et al. 2011; Taher et al. 2011; May et al. 2012; Arnold et al. 2014; Villar et al. 2015; Wong et al. 2020; Vaishnav et al. 2022). This suggests that there is considerable flexibility within enhancer sequences, and that the maintenance of function-defining features rather than overall sequence similarity is important for enhancer activity. This is illustrated most clearly by the maintenance of TF motifs at invariant positions or at different relative positions within orthologous enhancer sequences (Ludwig et al. 1998, 2000; Rastegar et al. 2008; Arnold et al. 2014; Wong et al. 2020). However, how flexible or constrained motif positions within enhancers are at both, the DNA sequence and the TF motif level, that is, how many different sequence variants or motif types might functionally replace the wild-type sequence at important motif positions, has remained unknown. Similarly, even though TF motifs have been observed to move between different enhancer positions over the course of evolution (presumably a consequence of motif decay and de novo formation), and despite position independence being a key assumption of the billboard model, the influence of the position and sequence context on a motif's contribution to enhancer function is not understood. These knowledge gaps restrict our understanding of the functional and evolutionary flexibility of enhancer sequences and how many sequence variants, as they might arise by DNA mutagenesis, might lead to similar or different enhancer activities.
Here, we investigated how many defined DNA sequences might functionally replace the wild-type sequence in various motif and control positions by exhaustively testing all possible 8-nucleotide-long sequence variants at these positions in two enhancers in Drosophila melanogaster S2 cells. In addition, we systematically compared the contribution of prominent TF motif types to enhancer activity when placed into different positions along an enhancer to assess how their intrinsic strengths are modulated by the sequence context in both Drosophila as well as human enhancers. Overall, these complementary approaches emphasize the flexibility of enhancer sequences and how the activity of TF motifs is modulated by the enhancer sequence context, namely the flanking sequence, the presence and diversity of other motif types, and the distance between motifs.
Results
STARR-seq comprehensively assesses the activity of enhancer variants revealing constrained enhancer sequence flexibility
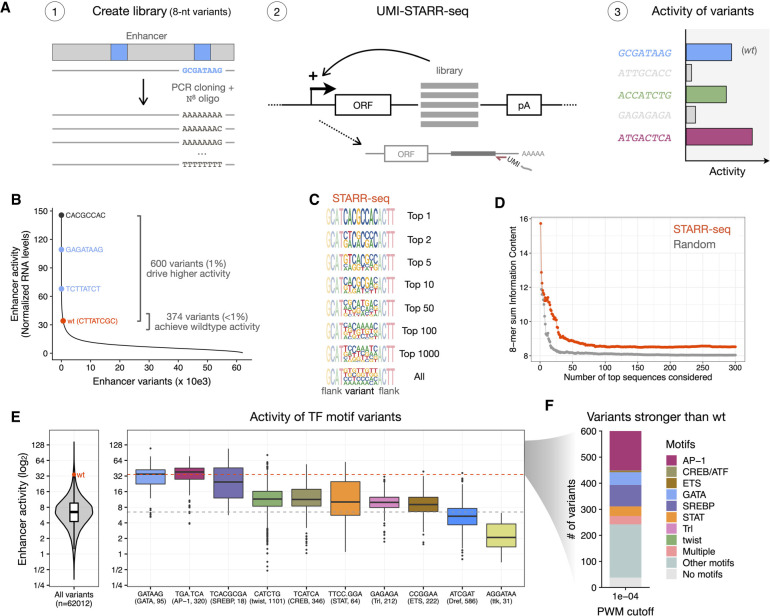
To systematically test what sequences function in a certain enhancer position, we used an approach inspired by studies that tested the activity of fully randomized regulatory sequences (Farley et al. 2015; de Boer et al. 2020; Vaishnav et al. 2022; Galupa et al. 2023) or the local fitness landscape of the green fluorescent protein (GFP; Sarkisyan et al. 2016; Somermeyer et al. 2022). We generated a comprehensive library of sequence variants by replacing a specific 8-nt stretch in an enhancer with randomized nucleotides (N8) and assessed the enhancer activity of each variant by UMI-STARR-seq in Drosophila S2 cells (Fig. 1A; see Methods; Arnold et al. 2013; Neumayr et al. 2019). Enhancer activity as used here is a quantitative measure and is defined as the increase in transcription of the reporter by a given enhancer candidate. We tested the power of this approach in the position of a GATA TF motif within the ced-6 developmental enhancer (ced-6 position 241 nt, or pos241) that is required for its activity. We recovered all possible 8-nt variants (65,536) in the input library and obtained reliable enhancer activity measurements for each variant (Supplemental Fig. S1). This showed that the vast majority of all variants drive low activity levels, whereas only 374 (<1%) achieve similar activity to wild type (±10%) and 600 (1%) drive even higher activity, that is, constitute valid solutions at this motif position (Fig. 1B).
Figure 1.
STARR-seq comprehensively assesses the activity of random variants in a specific enhancer position. (A) Schematics of STARR-seq for the analysis of random variants in an enhancer position: (1) A comprehensive library of sequence variants was generated by replacing the 8-nt stretch overlapping a GATA TF motif in the strong ced-6 enhancer with all possible 65,536 randomized nucleotides; (2) the enhancer activity of each variant was measured by STARR-seq in Drosophila S2 cells; (3) expected outcomes include the wild-type sequence (wt, blue), inactive variants (gray), and variants that recover the wild-type activity (green) or are even stronger (purple). (B) Most sequence variants exhibit low activity levels. The distribution of enhancer activity for each of the 62,012 enhancer variants with confident activity is shown. The wild-type (wt, red) sequence, the strongest GATA variant in each orientation (blue), and the strongest sequence variant are highlighted, together with the number of variants that achieve similar activity to wild type (±10%) or drive even higher activity. (C) Strong sequence variants are highly diverse. Logos with nucleotide frequency of the most-active variants in STARR-seq (1, 2, 5, 10, 50, 100, 1000, and all) and flanking nucleotides. Please note that because variants are aligned this will smear out motifs that occur at different positions. Motif finding with HOMER for these variants is shown in Supplemental Figure S2. (D) Sum of information content within the most-active 8-mers in STARR-seq (red) compared with the same after randomly sorting the variants (gray), considering different number of top sequences. (E) Distribution of enhancer activity for all 62,012 enhancer variants (left) or variants creating each TF motif (right). The activity of the wild-type sequence (wt, red dot and dashed line) or median of all variants (gray dashed line) are shown. The string of each TF motif used for the motif matching and the number of variants matching to each motif are described in the x-axis in the format “motif string (TF motif name, number of variants).” (F) Number of variants among the 600 stronger than wild type that match to motifs enriched in S2 developmental enhancers (PWM P-value cutoff 1×10−4).
Although only a few hundred sequences functioned at this position, these were highly diverse (Fig. 1C,D) and included not only different variants of the GATA motif (Fig. 1B—in blue, and 1E,F) but also other TF motifs, such as SREBP and AP-1 (Fig. 1E,F; Supplemental Figs. S2A,B, S3A). The different levels of importance of motifs were independent of their orientation, with the possible exception of SREBP and STAT for which differences are apparent yet not significant and cannot be assessed reliably because of a small number of instances (Supplemental Fig. S3A). Most of the 600 variants stronger than wild type (94%) created TF motifs overrepresented in S2 developmental enhancers (PWM P-value 1×10−4; Fig. 1F; Supplemental Fig. S3B), showing that there is flexibility in the DNA sequences but also in the motif types they encode. However, different TF motifs rescued enhancer activity to different levels (Fig. 1E; Supplemental Fig. S3A). Whereas AP-1 and SREBP achieved similar activity to the wild-type GATA motif, twist and ETS had lower activity at this enhancer position, despite being generally associated with strong enhancer activity in S2 cells (de Almeida et al. 2022). Therefore, the observed sequence flexibility is constrained to some TF motifs. In addition, even within each TF motif not all specific sequence variants functioned similarly, as apparent in the large differences between their activities (Fig. 1E). We observed a positive association between the activities of motif sequence variants and the TF motif affinities for most motifs, yet the correlation was typically modest, indicating that the PWM motif score does not explain the widely different activities (only SREBP has a PCC > 0.6 and twist and ETS even have PCCs < 0.1; Supplemental Fig. S3C).
We also observed TF motif types that had neutral or repressive functions at the tested 8-nt position: The Dref motif, previously shown to only be important for housekeeping enhancers (Zabidi et al. 2015; de Almeida et al. 2022), had no activity in this ced-6 developmental enhancer, whereas the Ttk motif created the most inactive 8-nt variants consistent with Ttk's function as a repressor (Fig. 1E; Supplemental Fig. S2C; Xiong and Montell 1993). These results show that this approach can comprehensively assess the activity of all sequence variants in a specific region of the enhancer and identify activating, neutral, and repressive sequences. Moreover, our findings indicate that developmental enhancers exhibit constrained flexibility, in that many variants, but still a strongly restricted number, can function at a given enhancer position. This constrained sequence flexibility applies not only to individual DNA sequences but also TF motif types in that several different motif types work, but not many or all.
Activity of random variants in seven specific positions of two different enhancers
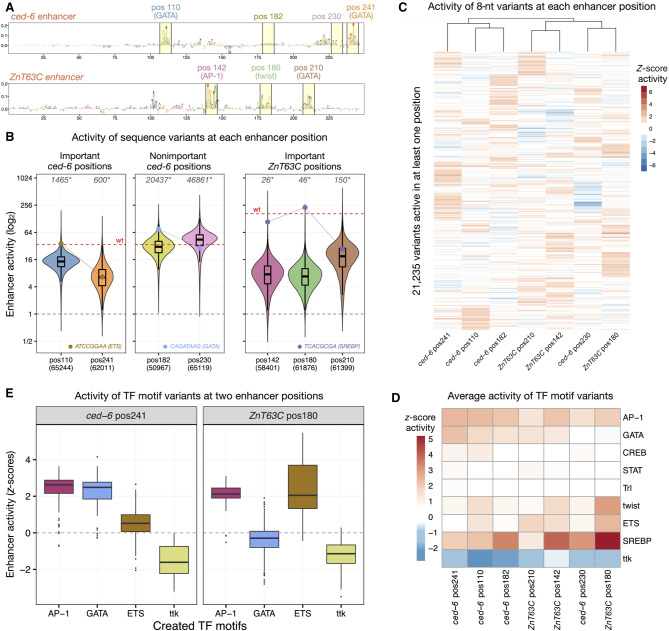
To evaluate if the same principles and the same specific solutions apply at different enhancer positions, we selected three additional positions of the ced-6 enhancer and three positions of a strong enhancer in the ZnT63C locus (Fig. 2A). To probe enhancer sequence flexibility at important motif positions and nonimportant control positions, we used the deep learning model DeepSTARR (Fig. 2A; de Almeida et al. 2022) and previous experimental enhancer mutations (Supplemental Fig. S4F) to choose positions that should (ced-6 pos110, pos241; ZnT63C pos142, pos180, pos210) or should not (ced-6 pos182, pos230) be important for enhancer activity. We generated exhaustive libraries of all 8-nt sequence variants for each position and performed UMI-STARR-seq on the combined libraries of each enhancer (Supplemental Fig. S4A–E; see Methods). As observed for the GATA position in Figure 1 (pos241), only a restricted set of variants achieved wild-type activity at a second important GATA motif position in the same enhancer (pos110) or at the important motif positions in the ZnT63C enhancer (Fig. 2B), confirming that important positions in enhancers show constrained flexibility. This contrasted with the nonimportant positions (pos182 and pos230 of the ced-6 enhancer) where most sequence variants were active at or near wild-type levels (Fig. 2B). This is expected, as these positions are predicted to not contain sequences associated with enhancer activity and are therefore less constrained. Thus, the importance of an enhancer position reflects its constraint, with nonimportant positions not being constrained (while they can still be modulated positively or negatively).
Figure 2.
Sequence constraints at different enhancer positions. (A) DeepSTARR-predicted nucleotide contribution scores for the ced-6 (top) and ZnT63C (bottom) selected enhancer sequences. Selected 8-nt motif positions and nonimportant control positions are highlighted in yellow with the respective numerical position, TF motif identity, and different colors. (B) Distribution of enhancer activity for all enhancer variants detected in each enhancer position. The activity of the wild-type sequence of each enhancer (wt, red dashed line) or of inactive sequences (gray dashed line) are highlighted, together with the activity of example sequence variants that create different TF motifs (ETS, GATA, and SREBP; dots and connected lines). Number of variants tested in each position are shown on the x-axis, whereas the number of variants with higher activity than wild type is shown on the top (gray, *). (C) Heatmap of Z-scores of log2 enhancer activity of 21,235 variants across all seven enhancer positions. Only variants assessed in all positions and active (Z-score > 1) in at least one are shown. Variants were clustered using hierarchical clustering and their activity is colored in shades of red (activating) and blue (repressing). (D) Heatmap of average Z-scores of log2 enhancer activity of variants creating each TF motif type (y-axis) across all enhancer positions (x-axis; sorted as in C). Motif activity is colored in shades of red (activating) and blue (repressing). (E) Distribution of Z-scores of log2 enhancer activity for variants creating each of four TF motifs (AP-1, GATA, ETS, ttk) in two selected enhancer positions (ced-6 pos241 and ZnT63C pos180).
The most active sequences at each enhancer position were highly diverse and exhibited distinct nucleotide preferences (Supplemental Figs. S5–S7). For example, two positions located either in the ced-6 (pos110) or the ZnT63C (pos210) enhancer showed distinct preferences among the strongest 100 variants, which preferentially match to an SREBP (GTCAC[flanked by GTC]) or an ETS motif (CCGGA[A]), respectively (Supplemental Fig. S5B). These results show that different enhancer positions require different motif types and thus are under different constraints.
Different TF motif types are active at different enhancer positions
Comparing the activity of the 8-nt sequence variants between the enhancer positions (scaled to the average activity of variants to be comparable across positions; see Methods) revealed that they indeed functioned differently at different positions (Pearson correlation coefficients [PCCs] below 0.4 between positions; Fig. 2C; Supplemental Fig. S8A–C). Further consolidating the 8-nt into 6-nt variants to reduce the impact of the surrounding sequence of each position (averaged activity across the flanking nucleotides) showed higher correlations but still strong differences between positions (Supplemental Fig. S8A,B,D). The top variants and solutions of each position differed substantially, with each position revealing specific sequences with particularly high activity, matching to known TF motifs (Fig. 2C). For example, an ETS motif variant was among the strongest sequences at ced-6 pos110 but not at pos241, a GATA variant was very active at ced-6 pos182 but inactive at pos230, and an SREBP variant was active in all positions of the ZnT63C enhancer except at pos210 (Fig. 2B).
We next compared the activity of motifs between the seven positions of the two different enhancers, by consolidating the activity of all 8-nt variants (±4 nt flanks) creating each motif (Fig. 2D,E; Supplemental Fig. S9; see Methods). For each position the wild-type sequence as well as different variants of that motif were among the top variants. Whereas the repressor Ttk motif repressed in all positions and showed little specificity (similar to other known and novel repressor motifs; Supplemental Fig. S10), the activator motifs showed distinct profiles, such as motifs that are globally active in all positions (AP-1), motifs with low activity in all tested positions (STAT, CREB, and Trl), and motifs with highly context-dependent activities (GATA, twist, ETS, and SREBP) (Fig. 2D,E). For example, GATA was active at the ced-6 pos110 but not at the ZnT63C pos180 position, whereas ETS motifs showed the opposite profile with the strongest activity at ZnT63C pos180 (Fig. 2E). For GATA motifs, we observed strong activity in all positions except on ced-6 pos230 and ZnT63C pos180, which are positioned close to another GATA motif (Fig. 2A). This observation is in line with the previously observed negative interaction of GATA/GATA motif pairs at short distances (de Almeida et al. 2022) and suggests that the observed different activities of TF motifs at different enhancer positions depend on their interaction with other TFs and the sequence context.
In summary, testing thousands of sequence variants in different enhancer positions revealed that enhancer sequences display constrained flexibility, in that only a specific but still diverse set of sequences and TF motifs can function at a given position. However, these constraints and solutions differed between enhancer positions, with different TF motifs active at different positions, suggesting that their activity is modulated by the sequence context.
Systematic motif pasting shows that motifs work differently at different enhancer positions
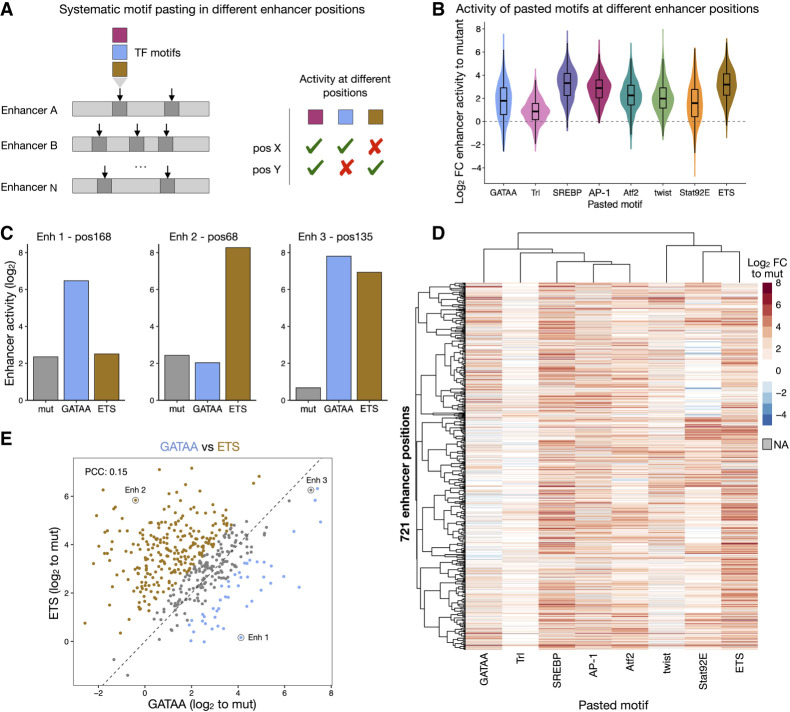
To systematically test if and how the enhancer sequence context modulates the function of TF motifs, we selected eight TF motifs that showed distinct position-dependent preferences (GATA, Trl, SREBP, AP-1, Atf2, twist, Stat92E, and ETS) and pasted their optimal sequences into 763 positions in a total of 496 developmental enhancers (Fig. 3A; see Methods). These positions were selected to be TF motifs important for the activity of the respective enhancers, as assessed by motif mutagenesis, allowing the reliable measurement of the increase in enhancer activity after pasting each TF motif (here quantified as the log2 fold-change activity over the motif-mutated enhancer). UMI-STARR-seq experiments with these designed libraries produced highly reproducible and quantitative enhancer activity measurements (replicates PCC between 0.94 and 0.98; Supplemental Fig. S11). Disrupting the selected enhancer positions by shuffling the wild-type sequences substantially reduced the activity of the respective wild-type enhancers by an average of more than sixfold, and pasting the different TF motifs in these same positions rescued enhancer activity to different levels (Supplemental Fig. S12A). Because we pasted the same optimal sequence for each TF motif into all positions, the differences in activity can only be explained by their respective sequence context; the differences between TF motifs are also directly comparable, because we pasted them in the same set of positions.
Figure 3.
TF motifs work differently at different enhancer positions. (A) Schematics of systematic motif pasting in different enhancer positions. Eight TF motifs that showed distinct position-dependent preferences were selected and their optimal sequence was pasted in 763 positions distributed among 496 enhancers, representing different contexts. The enhancer activity of each variant was measured by STARR-seq in Drosophila S2 cells to quantify the activity of motifs at the different positions. (B) Distribution of enhancer activity changes (log2 FC to mutated sequence) across all enhancer positions for each pasted TF motif. (C) Bar plots with activity (log2) of variants of three different enhancers with a mutated sequence (gray), a GATA (blue), or a ETS (brown) motif pasted at the same position. (D) Heatmap of enhancer activity changes (log2 FC to mutated sequence) after pasting each of the eight selected TF motifs in 721 enhancer positions (positions with data for at least six motifs). TF motifs and positions were clustered using hierarchical clustering and the activity is colored in shades of red (activating) and blue (repressing); missing values are colored in gray. (E) GATA and ETS motifs work differently at different enhancer positions. Comparison between enhancer activity changes (log2 FC to mutated sequence) after pasting GATA (x-axis) or ETS (y-axis) across all enhancer positions. Positions with stronger activity of GATA or ETS (≥twofold with respect to the other motif) are colored in blue and brown, respectively. Enhancer positions shown in C are highlighted. PCC: Pearson correlation coefficient.
Across all positions TF motifs had different median activities, which we interpret as different intrinsic strengths, with SREBP, ETS, and AP-1 being the strongest motifs and Trl the weakest (Fig. 3B; Supplemental Fig. S12A). However, enhancer positions had large effects on the motif activities that differed more than 100-fold for the same motif (Fig. 3B). For example, pasting a GATA motif activated enhancer activity more than 20-fold for 33 positions but not at all for 72 different positions. This position dependency was particularly strong for Trl, Stat92E, and GATA motifs, and weaker for AP-1, SREBP, and ETS (Supplemental Fig. S12B), which all had higher intrinsic strengths. Additionally, each TF motif showed differential activity across enhancer positions and activated in a unique set of positions. For example (Fig. 3C), GATA motifs activated enhancer1-position168 but not enh2-pos68, whereas ETS showed the opposite effect, and both motifs activated enh3-pos135. The different TF motifs showed different activity profiles across all positions, as revealed by global comparisons and hierarchical clustering (Fig. 3D; Supplemental Fig. S13). These results highlight the complexity of enhancer syntax and the difficulty of predicting and interpreting individual sequence manipulations.
The distinct preferences observed between pasted motifs were largely independent of the identity of the replaced wild-type motif across all positions, as revealed by the weak interaction scores between the wild type and the pasted motif identity in a multivariate linear regression analysis of all motif-pasting experiments (<1% explained variance, Supplemental Fig. S14). In contrast, the pasted motif identity (irrespective of the identity of the replaced motif) explains the most (23%) whereas 65% of variance remains unexplained and is likely due to surrounding enhancer sequence features affecting the motifs’ activities. Thus, systematic pasting of TF motifs across hundreds of enhancer contexts shows that motifs have different intrinsic strengths but work differently at different enhancers and positions, suggesting that the enhancer sequence context constrains the activity of TF motifs.
TF motifs have different intrinsic strengths that are modulated by the enhancer sequence context
The observed differential activities of motifs in different enhancer positions (Fig. 3D) suggest that the enhancer sequence context modulates the function of TF motifs. We found no significant differences when comparing the motif activity between pairs of positions in the same enhancer or in different enhancers, suggesting that the local context immediately surrounding the motif is as important as enhancer identity (Supplemental Fig. S15).
More globally, the sequence context for a motif can be related to its position within the enhancer, the motif flanking sequence, and the presence and distance to other motifs. To characterize the importance of these features, we tested if they contribute to the performance of predicting enhancer activity following the pasting of a motif at different enhancer positions. We first built a baseline random forest model that only includes the importance of the wild-type motif and the identity of the wild-type and pasted motifs as features, thereby not taking any sequence context features into account. This model obtained a PCC of 0.59 in the whole data set using tenfold cross-validation and showed that the pasted motif and the wild-type motif importance are strong determinants for enhancer activity (Supplemental Fig. S16A). Training a second random forest model that also includes context features such as the motif position relative to the enhancer center, the motif flanking sequence (defined as ±5 bp around the optimal motif as in de Almeida et al. [2022]), and the presence and distance to other TF motifs, improved this performance to a PCC of 0.69 (Supplemental Fig. S16B). This shows that the enhancer sequence context, particularly the closest flanking nucleotides as well as the presence of other motifs at specific distances (e.g., GATA or ETS), has an impact on the activity of TF motifs (Supplemental Fig. S16B).
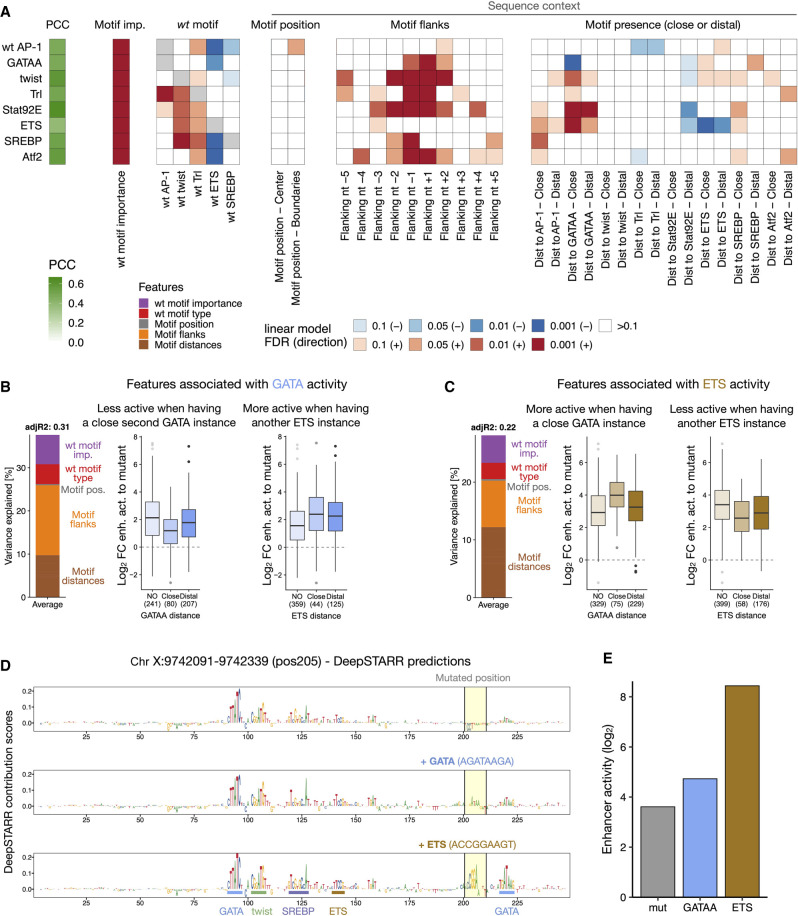
To better characterize the importance of these sequence rules for each TF motif separately, we generated interpretable linear models based on these rules to predict the motif activities across all positions (Fig. 4A). These models were able to predict the motif pasting results, with PCCs to experimentally assessed log2 fold changes between 0.39 (ETS) and 0.64 (Stat92E) (Fig. 4A; Supplemental Fig. S17). The motif flanks and the presence of additional motifs explained on average 16.7% and 6.7% of the motif activities variance, respectively, whereas the motif position within the enhancer had lower importance (0.4%).
Figure 4.
Characterization of preferred syntax features of each TF motif. (A) Motif syntax rules modulate TF motif function. For each TF motif type (rows), a linear model was built to predict its activity across all enhancer positions, using as covariates the number of instances, the wild-type TF motif importance and identity, and sequence context features such as the position within the enhancer, the flanking nucleotides, and the presence at close or distal distances to all other TF motifs. The PCC between predicted and observed motif activities is shown with the green color scale on the left. The heatmap shows the contribution of each feature (columns) for each model, colored by the FDR-corrected P-value (red or blue scale depending on positive or negative association, respectively). (B,C) Syntax features associated with GATA (B) or ETS (C) activity. Left: bar plot showing the variance explained by the different types of features (color legend) for each of the linear models. Middle and right: enhancer activity changes (log2 FC to a mutated sequence) after pasting each TF motif in positions with no additional GATA (middle) or ETS (right) in the enhancer, or with additional GATA or ETS at close (≤25 bp) or distal (>25 bp) distances. Number of instances are shown. (D) DeepSTARR-predicted importance scores for pasting a mutant sequence (gray), GATA (blue), or ETS (brown) in a specific position (Chr X: 9,742,091–9,742,339, pos205). Motif sequences pasted are shown. (E) Bar plots with measured enhancer activity (log2) of variants from D.
The TF motif type-specific models revealed how the sequence context rules differ between TF motif types, explaining the motif-specific enhancer position preferences. For example, GATA activity was strongly dependent on the flanking nucleotides and was modulated by the presence of a second GATA at close distance (negative interaction) or ETS motifs (positive interaction) (Fig. 4B; Supplemental Fig. S18A). We saw different associations for ETS activity, as expected by the different GATA and ETS activity profiles across all positions (Fig. 3E). ETS activity was only mildly influenced by the flanking nucleotides but strongly by neighboring motifs: it was stronger close to GATA motifs and weaker in enhancers with another ETS motif (Fig. 4C; Supplemental Fig. S18B). These sequence features, such as the negative GATA/GATA and the positive ETS/GATA interactions at close distances, were observed previously via computational models of wild-type S2 enhancer sequences (de Almeida et al. 2022).
In addition, the DeepSTARR-predicted importance of each nucleotide when pasting different TF motifs into the same position revealed their interaction with the sequence context (Fig. 4D,E; Supplemental Fig. S19): GATA but not ETS activated the Chr 3L enhancer in a position with additional distal GATA motifs, while ETS but not GATA activated the Chr X enhancer in a position with a GATA motif at close distance, and both activated the Chr 2L enhancer that contains multiple surrounding twist motifs, all consistent with these motifs’ respective distance preferences (de Almeida et al. 2022). Together these results demonstrate how the sequence context (e.g., the flanking sequence, and the presence and diversity of other motif types) modulates the function of TF motifs, constraining enhancer sequence flexibility.
Enhancer sequence context modulates the function of human TF motifs
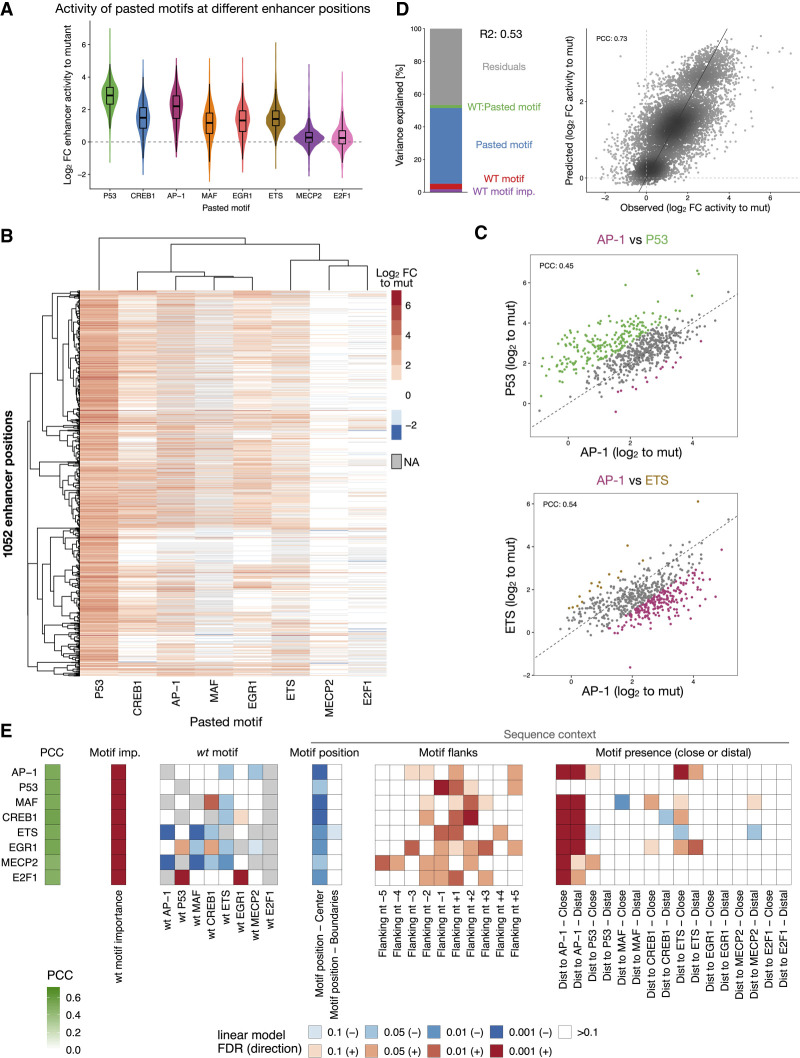
To test whether TF motifs also work differently in different enhancer sequence contexts in other species, we performed the systematic motif pasting experiment in human HCT116 cells for eight previously characterized human TF motifs (P53, AP-1, ETS, CREB1, MAF, EGR1, E2F1 and MECP2; see Methods; de Almeida et al. 2022). Pasting of the motifs into 1354 important positions in 753 different HCT116 enhancers revealed that human TF motifs also have different intrinsic strengths and work differently in different enhancers and positions (Fig. 5A; Supplemental Figs. S20, S21). P53 was the strongest motif and the only one that showed globally strong activity across all enhancer positions, suggesting little dependence on the enhancer context, as has been suggested before (Verfaillie et al. 2016). AP-1, the second strongest motif, was strongly dependent on the enhancer positions, with activities ranging more than 50-fold across enhancer contexts. This position dependence was also observed for the other motifs, even though their overall activity was lower (Fig. 5A).
Figure 5.
Human TF motifs require specific enhancer sequence contexts. (A) Distribution of enhancer activity changes (log2 FC to mutated sequence) across all enhancer positions for each pasted TF motif. (B) Heatmap of enhancer activity changes (log2 FC to mutated sequence) after pasting each of the eight selected human TF motifs in 1052 enhancer positions (positions with data for at least six motifs). TF motifs and positions were clustered using hierarchical clustering and the activity is colored in shades of red (activating) and blue (repressing); missing values are colored in gray. (C) Human TF motifs work differently at different enhancer positions. Comparison between enhancer activity changes (log2 FC to mutated sequence) after pasting AP-1 (x-axis) and P53 (top) or ETS (bottom) (y-axis), across all enhancer positions. Positions with stronger activity of each motif (≥twofold in respect to the other motif in the scatter plot) are colored (P53: green, AP-1: purple, ETS: brown). PCC: Pearson correlation coefficient. (D) TF motif activity in function of wild-type and pasted motif identity. Left: Bar plot showing the amount of variance explained by the wild-type motif importance and identity, the pasted motif identity, and the interaction between the wild-type and pasted motifs, using a linear model fit on all motif pasting results. Right: Scatter plots of predicted (linear model) versus observed enhancer activity changes (log2 FC to mutated sequence) across all motif pasting experiments. Color reflects point density. (E) Motif syntax rules modulate the function of human TF motifs. For each TF motif type (rows), we built a linear model to predict their activity across all enhancer positions, using as covariates the number of instances, the wild-type TF motif importance and identity, and sequence context features such as the position within the enhancer, the flanking nucleotides, and the presence at close or distal distances to all other TF motifs. The PCC between predicted and observed motif activities is shown with the green color scale on the left. The heatmap shows the contribution of each feature (columns) for each model, colored by the FDR-corrected P-value (red or blue scale depending on positive or negative association, respectively).
TF motifs preferred different enhancer contexts, with four groups of motifs showing characteristically different preferences: (1 – P53) strong activity in all positions; (2 – CREB1, AP-1, MAF, EGR1) and (3 – ETS) highly context-dependent activities; (4 – MECP2, E2F1) only active in few and highly specific enhancer positions (Fig. 5B,C; Supplemental Fig. S22). These distinct preferences were independent of the identity of the replaced motif (Fig. 5D; Supplemental Fig. S23) but correlated with sequence context features. Similar to Drosophila TF motifs, motif context features such as motif flanks and the presence and distance to other TF motifs were important to predict the activities of human motifs across the different enhancer positions (Supplemental Fig. S24). TF-specific linear models based on such syntax features were able to predict the motif activities across all positions (PCCs between 0.46 and 0.51; Supplemental Fig. S25) and revealed the context preferences of each TF motif (Fig. 5E).
All motif activities were influenced by the flanking nucleotides, which explained on average 8.2% of the motif activities' variance, whereas the presence of additional motifs and their distance explained 8.5% (Fig. 5E; Supplemental Figs. S25, S26). As expected by the weak context specificity of P53 (group 1, Fig. 5A), its activity was independent of the presence and distance to other TF motifs (Fig. 5E; Supplemental Fig. S26A). All the other motifs preferred contexts with an additional AP-1 instance (Fig. 5E). The AP-1 motif itself, as well as MAF, CREB1, and EGR1 (group 2), all preferred positions close to an ETS motif, concordant with previous studies showing direct protein–protein interactions between ETS and other TFs (Li et al. 2000; Burda et al. 2010), whereas the ETS motif (group 3) had a negative interaction with a second close ETS motif (Fig. 5E), as also observed in Drosophila enhancers (Fig. 4A). These findings are also concordant with the motif syntax rules found in a previous study (de Almeida et al. 2022). Altogether, this establishes that TF motifs require specific enhancer sequence contexts in species as divergent as fly and human, suggesting that this is a general principle of regulatory enhancer sequences.
Discussion
In this study, we used two complementary strategies to explore the flexibility of enhancers with regard to nucleotide and motif identity at specific enhancer positions as well as the position dependence of motif activity. Even though median enhancer activity drops significantly when randomizing an 8-nt stretch at important positions, many sequence variants, including variants of the wild-type motif but also other TF motifs, can achieve strong enhancer activity. The diverse solutions at each position show that enhancers exhibit some degree of flexibility. However, as only a few hundred out of the >65,000 tested sequences work, the flexibility at any given position is constrained. Similarly, systematically pasting different motifs into hundreds of enhancer positions revealed that motif activity is strongly modulated by the enhancer sequence context. Therefore, constrained sequence flexibility and the modulation of motif function by the sequence context seem to be key features of enhancers.
The observation that both Drosophila and human TF motifs require specific enhancer sequence contexts suggests that this is a general principle of enhancers. Even though motifs possess some intrinsic strengths, their potential to activate transcription strongly depends on the sequence context and follows certain syntax rules, including motif flanks, combinations, and distances. Although our study cannot assess the mechanistic causes for these rules, they might be related to local DNA shape (Dror et al. 2015; Mathelier et al. 2016; Samee et al. 2019) or to more general enhancer DNA properties such as DNA bending. Our observation that homotypic interactions of certain motifs at close distances (e.g., GATA or ETS) are negatively associated with enhancer activity is consistent with repressive homotypic interactions between pluripotency TFs found by thermodynamic modeling (Fiore and Cohen 2016); the mechanisms, however, are still unclear. Intermotif distances can impact the synergy between TFs at the level of DNA binding or after binding, such as cofactor recruitment and activation, which could explain both positive and negative TF-TF interactions (Reiter et al. 2017). Although these syntax rules seem to be stricter for some TF motifs (e.g., GATA) and more relaxed for others (e.g., P53), our results show that motifs are not simply independent modules. Instead, they interact with all enhancer features in a highly cooperative manner, which can modulate motif activity by more than 100-fold. This is an important result that supports a model where enhancer activity is encoded through a complex interdependence between motifs and context, rather than motifs acting independently and additively. Whereas tissue- or cell type–specificity can already be predicted by motif presence-absence patterns alone (Kvon et al. 2014; Janssens et al. 2022), the encoding of different enhancer strengths seems to depend on more complex cis-regulatory syntax rules (Jindal and Farley 2021; de Almeida et al. 2022). The functional implications of mutations in TF motifs or elsewhere within enhancer sequences can therefore only be assessed in the context of these syntax features.
The motif syntax rules described here agree well with the ones learned by DeepSTARR trained on genome-wide enhancer activity data (de Almeida et al. 2022) and the BPNet model trained on endogenous TF binding and cooperativity (Avsec et al. 2021), suggesting that these rules are important in wild-type enhancer sequences. As an ectopic reporter assay STARR-seq measures the potential of sequences to act as enhancers, even if the sequences might be repressed endogenously at the chromatin level (Arnold et al. 2013; Muerdter et al. 2018), making it a powerful tool to uncover the sequence determinants for enhancer activity. It will be interesting to explore the sequence rules and mechanisms by which chromatin modulates endogenous enhancer activities and gene expression using complementary methods (Catarino and Stark 2018). In addition, DeepSTARR also predicted with good accuracy the activity of all randomized sequence variants and of motifs pasted in different enhancer contexts (Supplemental Figs. S27, S28). This supports the validity of computational models such as DeepSTARR and their use in in-silico-like experiments (e.g., motif pasting experiments with a larger set of TF motifs across many more genomic positions) to improve our understanding of the regulatory information encoded in enhancer sequences and the impact of mutations.
Our study shows that enhancer sequences are flexible enough for enhancer strength to be achieved by a small yet diverse set of sequence variants, and that mutations in information-poor positions have little impact on the enhancer activity in a single cell type. This flexibility allows many different sequences to achieve similar enhancer activities in a single cell type, which might be an important prerequisite for the evolution of developmental enhancers that operate under many additional constraints, for example, regarding the precise spatiotemporal control of enhancer activities. As the activity in a given cell can be achieved by many solutions, the specific solutions that fulfill additional requirements can be explored during evolution. Indeed, previous studies that have analyzed expression changes of enhancer mutations across different cell types in vivo have observed that the cell type–specific expression patterns of enhancers can change upon (minimal) sequence perturbations (Farley et al. 2015; Fuqua et al. 2020; Galupa et al. 2023). The fact that enhancer strength in any given cell type and enhancer specificity across cell types and developmental time are subject to different yet overlapping sequence constraints highlights the complexity of the regulatory code. We expect that the combination of quantitative enhancer-sequence-to-function models in individual cell types and qualitative predictions of enhancer activities across cell types will provide unprecedented progress in our understanding of enhancer biology and our ability to read and write enhancer sequences.
Methods
UMI-STARR-seq library cloning
Libraries of Drosophila enhancer variants with 8-nt randomized sequences were generated using a PCR approach with degenerate oligonucleotides. Forward primers (Supplemental Table S1) were designed to anneal directly downstream of the enhancer position of interest followed by 8 degenerate bp (creating 65,536 variants) and another 20 bp complementary stretch. Reverse primers were complementary to the 20 bp 5′ of the degenerate stretch. The STARR-seq vector containing the wild-type enhancer of interest (ced-6 or ZnT63C) was used as a template for the PCR. The PCR was run across the whole STARR-seq plasmid, followed by DpnI digestion and a Gibson reaction that recircularizes the plasmid. Drosophila and human oligo libraries were amplified (for primers, see Supplemental Table S1) and cloned into Drosophila STARR-seq vectors containing the DSCP core promoter and into the human STARR-seq plasmid with the ORI in place of the core promoter (Muerdter et al. 2018), respectively. All libraries were grown in 2l LB-Amp (final ampicillin concentration 100 µg/mL). All libraries were purified with Qiagen Plasmid Plus Giga Kit (cat. no. 12991).
Cell culture, transfection, and UMI-STARR-seq
Drosophila S2 and human HCT116 cells were cultured as described previously (Arnold et al. 2013; Muerdter et al. 2018). Cells were electroporated using the MaxCyte-STX system at a density of 50 × 106 cells per 100 µL and 5 µg of DNA using the “Optimization 1” protocol (S2) and at a density of 1 × 107 cells per 100 µL and 20 µg of DNA using the preset “HCT116” program (HCT116), respectively. We transfected 400 × 106 S2 cells total per replicate with 20 μg of the input library for Drosophila and 80 × 106 HCT116 cells total per replicate with 160 µg of the input library for human cells. UMI-STARR-seq was performed as described previously (Arnold et al. 2013; Muerdter et al. 2018; Neumayr et al. 2019). Further experimental details can be found in the Supplemental Methods.
Random variant UMI-STARR-seq data analysis
RNA and DNA input reads (paired-end 150 bp) were mapped to dedicated Bowtie indices using Bowtie v.1.2.2 (Langmead et al. 2009). Because the N8 variants were all positioned in the last 150 nt of each enhancer, we allowed for flexible mapping in the beginning of the fragments to increase the number of mapped reads while keeping high sensitivity for the different enhancer variants. Specifically, we trimmed the forward reads to 36 bp and mapped them to the indices allowing for three mismatches; the full 150-bp-long reverse reads were mapped with no mismatches, to identify all sequence variants; paired-end reads with the correct position, length, and strand were kept. For paired-end DNA and RNA reads that mapped to the same variant, we collapsed those that have identical UMIs (10 bp, allowing one mismatch) to ensure the counting of unique molecules (Supplemental Table S2).
We excluded oligos with less than five reads in any of the input replicates and less than one read in any of the RNA replicates. The enhancer activity of each sequence in each screen was calculated as the log2 fold-change over input, using all replicates, with DESeq2 (Love et al. 2014).
Oligo library UMI-STARR-seq data analysis
As described previously (de Almeida et al. 2022), RNA and DNA input reads were mapped to a reference containing the 249-bp-long sequences from the fragments present in the Drosophila (dm3) or human (hg19) libraries using Bowtie v.1.2.2 (Langmead et al. 2009). We used these reference genomes to be able to integrate our results with older in-house and published data sets and made sure this choice does not affect the quantifications of enhancer activity. Mapping reads with the correct length, strand, and with no mismatches were kept. Both DNA and RNA reads were collapsed by UMIs (10 bp) as above (Supplemental Table S2).
We excluded oligos with less than 10 reads in any of the input replicates and added one read pseudocount to oligos with zero RNA counts. The enhancer activity of each oligo in each screen was calculated as the log2 fold-change over input, using all replicates, with DESeq2 (Love et al. 2014).
Random variant libraries of Drosophila enhancers and UMI-STARR-seq
Two strong S2 developmental enhancers with different TF motif compositions were selected to test a diversity of random 8-nt variants in different positions: ced-6 (Chr 2R: 5,326,628–5,326,876) and ZnT63C (Chr 3L: 3,310,914–3,311,162) enhancers. We selected five positions important for the activity of the two enhancers (ced-6 pos110 and pos241; ZnT63C pos142, pos180, pos210) and two nonimportant positions of the ced-6 enhancer (pos182 and pos230) and replaced each 8-nt stretch of the enhancer with randomized nucleotides (N8), creating 65,535 enhancer variants in addition to the wild-type sequence per position. For each enhancer, we pooled the libraries of the different positions and combined them with an oligo library of thousands of wild-type enhancers and negative sequences (de Almeida et al. 2022) for normalization. UMI-STARR-seq using the ced-6 or ZnT63C pooled libraries was performed and analyzed as described above (Supplemental Table S3). We performed two independent replicates per enhancer pooled library screen (Pearson correlation coefficient (PCC) = 0.85–0.91). To be able to compare the activity of variants and motifs between enhancer positions, we next scaled the enhancer activity of all variants per position (Z-scores). This allowed us to measure the change in activity of a given variant over the average of all variants, correcting for the importance of the different enhancer positions tested.
Diversity of top active variants and de novo motif discovery
The most-active 8-nt variants of each screen (1, 2, 5, 10, 50, 100, and 1000) were retrieved and consolidated into position probability matrices based on the nucleotide frequencies at each position. Logos were visualized using the ggseqlogo function from R package ggseqlogo (v.0.1; https://CRAN.R-project.org/package=ggseqlogo). The top 100 and 1000 or bottom 1000 variants (8 nt ± 4 nt flanks) of each screen were used for de novo motif discovery analyses using HOMER, taking all detected variants of the respective screen as background. HOMER (v4.10.4; Heinz et al. 2010) was run with the findMotifs.pl command and the arguments fly -len 6,7,8.
Activity of TF motifs created by sequence variants
To robustly assess the activity of a given TF motif, we retrieved the activity of all 16-nt variants (8 nt ± 4 nt flanks) creating each motif by string matching. For a more systematic comparison across all TF motif types, we matched variants to the optimal string from each TF motif PWM model in a motif database (de Almeida et al. 2022). The average activity across variants was defined as the motifs’ intrinsic strength. To find how many active variants are explained by the creation of known motifs enriched in S2 developmental enhancers, we performed PWM-based motif scanning of those candidate motifs onto variants (8 nt ± 4 nt flanks). We used the matchMotifs function from R package motifmatchr (v.1.4.0; genome = “BSgenome.Dmelanogaster.UCSC.dm3”, bg=“genome” [https://bioconductor.org/packages/release/bioc/html/motifmatchr.html]) with P-value cutoffs 1×10−4 and 1×10−5.
Comparison of random variants activity across enhancer positions
We compared the activity of all 8-nt random variants across enhancer positions using their Z-score scaled activity (Supplemental Table S3). We calculated pairwise PCCs between the different libraries, performed hierarchical clustering (“complete” method) using the correlation values as similarities, and displayed heatmaps using the pheatmap R package (v.1.0.12; https://CRAN.R-project.org/package=pheatmap). To reduce the impact of the flanking sequence of each position when comparing the activity of variants between them, we repeated the same after consolidating the 8-nt into shorter variants by taking the centered sequence and averaging the activity across variants with different flanking nucleotides.
Drosophila and human TF motif mutagenesis oligo library synthesis and UMI-STARR-seq
For the Drosophila library, we selected 1172 motif positions (among 728 enhancers) that are required for the activity of the respective enhancers and designed sequences of enhancer variants where we pasted a mutant sequence or the optimal sequence of eight TF motifs (GATA, AP-1, twist, Trl, ETS, SREBP, Stat92E, and Atf2; one at a time; sequences in Supplemental Table S4) in each of these positions. For the human library, we selected 1456 motif positions important for the activity of 808 enhancers and designed sequences of enhancer variants where we pasted a mutant sequence or the optimal sequence of the same eight TF motifs (AP-1, ETS, E2F1, EGR1, MAF, MECP2, CREB1, P53; one at a time; sequences in Supplemental Table S4) in each of these positions. Each of the Drosophila and human libraries was synthesized and pooled with a previous library containing the respective wild-type enhancer sequences (de Almeida et al. 2022) to be screened together (Supplemental Tables S5, S6). All details can be found in the Supplemental Methods. The resulting 300-mer oligonucleotide Drosophila and human libraries were synthesized by Twist Bioscience. UMI-STARR-seq using these oligo libraries was performed and analyzed as described above (Supplemental Tables S5, S6). We performed three independent replicates for Drosophila (correlation PCC = 0.95–0.98) and human (PCC = 0.96–0.98) screens.
Quantification of motif activity at different enhancer positions
We used our enhancer activity measures of the wild-type and mutated sequences to stringently select important enhancer positions for further analyses: positions where mutation reduced the activity by at least twofold (Supplemental Figs. S12A, S21A). These resulted in 763 important positions distributed among 496 Drosophila enhancers and 1354 positions distributed among 753 human enhancers. Variability of activity of each motif across enhancer positions was quantified using the coefficient of variation (ratio of the standard deviation to the mean; Supplemental Fig. S12B). We compared the activity of motifs across enhancer positions by pairwise PCCs and performed hierarchical clustering (“complete” method) using the correlation values as similarities. Heatmaps were displayed using the pheatmap R package (v.1.0.12; https://CRAN.R-project.org/package=pheatmap).
Prediction of motif activities using motif syntax features
We extracted the following syntax features per tested enhancer position: the position relative to the enhancer center (center: −/+25 bp, flanks: −/+25:75 bp, boundaries: −/+75:125 bp), the position flanking nucleotides (5 bp on each side), and the presence and distance to other TF motifs (close: ≤25 bp; distal: > 25 bp; between motif centers). Instances of each TF motif type were mapped across all enhancers using their annotated PWM models (Supplemental Table S3) and the matchMotifs function from R package motifmatchr (v.1.4.0; https://bioconductor.org/packages/release/bioc/html/motifmatchr.html) with the following parameters: genome = “BSgenome.Dmelanogaster.UCSC.dm3”, p.cutoff = 5e-04, bg = “genome”.
We used a 10-fold cross-validation scheme to train random forest models to predict Drosophila or human motif pasting activities (log2 fold-change to mutant) using as features the wild-type TF motif identity and importance (log2 fold-change activity between wild-type and motif-mutant sequence) and the pasted motif identity, together or not with the syntax features described above. All models were built using the caret R package (v. 6.0–80; https://CRAN.R-project.org/package=caret) and feature importance was calculated using its varImp function.
In addition, we trained a multiple linear regression model per TF motif type to predict its activity across different enhancer positions using as covariates the wild-type TF motif identity and importance together with the syntax features described above. All models were built using the caret R package (v. 6.0–80; https://CRAN.R-project.org/package=caret) and 10-fold cross-validation. The linear model coefficients and respective FDR-corrected P-values were used as metrics of importance for each feature, using the red or blue scale depending on positive or negative associations (Figs. 4A, 5E). We calculated the percentage of variance explained by each covariate in the linear models built for each TF motif with one-way ANOVAs. Further details can be found in the Supplemental Methods.
DeepSTARR nucleotide contribution scores and predictions of enhancer sequence changes
Nucleotide contribution scores for wild-type enhancers or enhancer variants were calculated using DeepSTARR as described previously (de Almeida et al. 2022) and visualized using the ggseqlogo function from the R package ggseqlogo (v.0.1; https://CRAN.R-project.org/package=ggseqlogo). DeepSTARR was also used to predict the enhancer activity of N8 variants in enhancers or the log2 fold-change enhancer activity of motif pasting sequences.
Statistics and data visualization
All statistical calculations and graphical displays have been performed in R statistical computing environment (v.3.5.1; R Core Team 2020) and using the R package ggplot2 (Wickham 2016). In all box plots, the central line denotes the median, the box encompasses 25th to 75th percentile (interquartile range), and the whiskers extend to 1.5× interquartile range.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE211659 or Zenodo (https://zenodo.org/record/7010528#.ZAeEay1h2v4). Code used to process the UMI-STARR-seq data as well as to reproduce all analyses, results, and figures has been submitted to GitHub (https://github.com/bernardo-de-almeida/Variant_STARRseq) and is available as Supplemental Code.
Supplementary Material
Acknowledgments
We thank V. Loubiere and T. Pachano (IMP) for comments on the manuscript and all members of the Stark group for discussions. Deep sequencing was performed at the Vienna Biocenter Core Facilities GmbH. F.R. is a recipient of a DOC Fellowship of the Austrian Academy of Sciences at the Research Institute of Molecular Pathology. Research in the Stark group is supported by the Austrian Science Fund (FWF). Basic research at the IMP is supported by Boehringer Ingelheim GmbH and the Austrian Research Promotion Agency (FFG).
Author contributions: F.R., B.P.d.A., and A.S. conceived the project. F.R. performed all experiments. B.P.d.A. performed all computational analyses. F.R., B.P.d.A., and A.S. interpreted the data and wrote the manuscript. A.S. supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277246.122.
Competing interest statement
The authors declare no competing interests.
References
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet 46: 685–692. 10.1038/ng.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898. 10.1002/jcb.20352 [DOI] [PubMed] [Google Scholar]
- Avsec Ž, Weilert M, Shrikumar A, Krueger S, Alexandari A, Dalal K, Fropf R, Mcanany C, Gagneur J, Kundaje A, et al. 2021. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat Genet 53: 354–366. 10.1038/s41588-021-00782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. 1981. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308. 10.1016/0092-8674(81)90413-X [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2010. ChIP-seq identification of weakly conserved heart enhancers. Nat Genet 42: 806–810. 10.1038/ng.650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Laslo P, Stopka T. 2010. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 24: 1249–1257. 10.1038/leu.2010.104 [DOI] [PubMed] [Google Scholar]
- Catarino RR, Stark A. 2018. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev 32: 202–223. 10.1101/gad.310367.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida BP, Reiter F, Pagani M, Stark A. 2022. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers. Nat Genet 54: 613–624. 10.1038/s41588-022-01048-5 [DOI] [PubMed] [Google Scholar]
- de Boer CG, Vaishnav ED, Sadeh R, Abeyta EL, Friedman N, Regev A. 2020. Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat Biotechnol 38: 56–65. 10.1038/s41587-019-0315-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. 2015. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res 25: 1268–1280. 10.1101/gr.184671.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. 2015. Suboptimization of developmental enhancers. Science 350: 325–328. 10.1126/science.aac6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C, Cohen BA. 2016. Interactions between pluripotency factors specify cis-regulation in embryonic stem cells. Genome Res 26: 778–786. 10.1101/gr.200733.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua T, Jordan J, van Breugel ME, Halavatyi A, Tischer C, Polidoro P, Abe N, Tsai A, Mann RS, Stern DL, et al. 2020. Dense and pleiotropic regulatory information in a developmental enhancer. Nature 587: 235–239. 10.1038/s41586-020-2816-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Alvarez-Canales G, Borst NO, Fuqua T, Gandara L, Misunou N, Richter K, Alves MRP, Karumbi E, Perkins ML, et al. 2023. Enhancer architecture and chromatin accessibility constrain phenotypic space during Drosophila development. Dev Cell 58: 51–62.e4. 10.1016/j.devcel.2022.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens J, Aibar S, Taskiran II, Ismail JN, Spanier KI, González-Blas CB, Quan XJ, Papasokrati D, Hulselmans G, Makhzami S, et al. 2022. Decoding gene regulation in the fly brain. Nature 601: 630–636. 10.1038/s41586-021-04262-z [DOI] [PubMed] [Google Scholar]
- Jindal GA, Farley EK. 2021. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev Cell 56: 575–587. 10.1016/j.devcel.2021.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Arnosti DN. 2003. Information display by transcriptional enhancers. Development 130: 6569–6575. 10.1242/dev.00890 [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A. 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. 10.1038/nature13395 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763. 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei H, Watson DK. 2000. Regulation of Ets function by protein–protein interactions. Oncogene 19: 6514–6523. 10.1038/sj.onc.1204035 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125: 949–958. 10.1242/dev.125.5.949 [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. 10.1038/35000615 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Xin B, Chiu TP, Yang L, Rohs R, Wasserman WW. 2016. DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst 3: 278–286.e4. 10.1016/j.cels.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. 2012. Large-scale discovery of enhancers from human heart tissue. Nat Genet 44: 89–93. 10.1038/ng.1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń ŁM, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, et al. 2018. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods 15: 141–149. 10.1038/nmeth.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayr C, Pagani M, Stark A, Arnold CD. 2019. STARR-seq and UMI-STARR-seq: assessing enhancer activities for genome-wide-, high-, and low-complexity candidate libraries. Curr Protoc Mol Biol 128: e105. 10.1002/cpmb.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D. 2008. The enhanceosome. Curr Opin Struct Biol 18: 236–242. 10.1016/j.sbi.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Rastegar S, Hess I, Dickmeis T, Nicod JC, Ertzer R, Hadzhiev Y, Thies WG, Scherer G, Strähle U. 2008. The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev Biol 318: 366–377. 10.1016/j.ydbio.2008.03.034 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reiter F, Wienerroither S, Stark A. 2017. Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev 43: 73–81. 10.1016/j.gde.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Rickels R, Shilatifard A. 2018. Enhancer logic and mechanics in development and disease. Trends Cell Biol 28: 608–630. 10.1016/j.tcb.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Samee MAH, Bruneau BG, Pollard KS. 2019. A de novo shape motif discovery algorithm reveals preferences of transcription factors for DNA shape beyond sequence motifs. Cell Syst 8: 27–42.e6. 10.1016/j.cels.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, et al. 2016. Local fitness landscape of the green fluorescent protein. Nature 533: 397–401. 10.1038/nature17995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. 10.1126/science.1186176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somermeyer LG, Fleiss A, Mishin AS, Bozhanova NG, Igolkina AA, Meiler J, Alaball Pujol M-E, Putintseva EV, Sarkisyan KS, Kondrashov FA. 2022. Heterogeneity of the GFP fitness landscape and data-driven protein design. eLife 11: e75842. 10.7554/eLife.75842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Swanson CI, Schwimmer DB, Barolo S. 2011. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr Biol 21: 1186–1196. 10.1016/j.cub.2011.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, Ovcharenko I. 2011. Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res 21: 1139–1149. 10.1101/gr.119016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. 1995. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83: 1091–1100. 10.1016/0092-8674(95)90136-1 [DOI] [PubMed] [Google Scholar]
- Vaishnav ED, de Boer CG, Molinet J, Yassour M, Fan L, Adiconis X, Thompson DA, Levin JZ, Cubillos FA, Regev A. 2022. The evolution, evolvability and engineering of gene regulatory DNA. Nature 603: 455–463. 10.1038/s41586-022-04506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie A, Svetlichnyy D, Imrichova H, Davie K, Fiers M, Atak ZK, Hulselmans G, Christiaens V, Aerts S. 2016. Multiplex enhancer-reporter assays uncover unsophisticated TP53 enhancer logic. Genome Res 26: 882–895. 10.1101/gr.204149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. 10.1016/j.cell.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. 2009. Genomic views of distant-acting enhancers. Nature 461: 199–205. 10.1038/nature08451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vockley CM, McDowell IC, D'Ippolito AM, Reddy TE. 2017. A long-range flexible billboard model of gene activation. Transcription 8: 261–267. 10.1080/21541264.2017.1317694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. 2010. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet 26: 66–74. 10.1016/j.tig.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. ISBN 978-3-319-24277-4, http://ggplot2.org. [Google Scholar]
- Wong ES, Zheng D, Tan SZ, Bower NI, Garside V, Vanwalleghem G, Gaiti F, Scott E, Hogan BM, Kikuchi K, et al. 2020. Deep conservation of the enhancer regulatory code in animals. Science 370: eaax8137. 10.1126/science.aax8137 [DOI] [PubMed] [Google Scholar]
- Xiong W-C, Montell C. 1993. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev 7: 1085–1096. 10.1101/gad.7.6.1085 [DOI] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. 2015. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. 10.1038/nature13994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.