FIGURE 2.
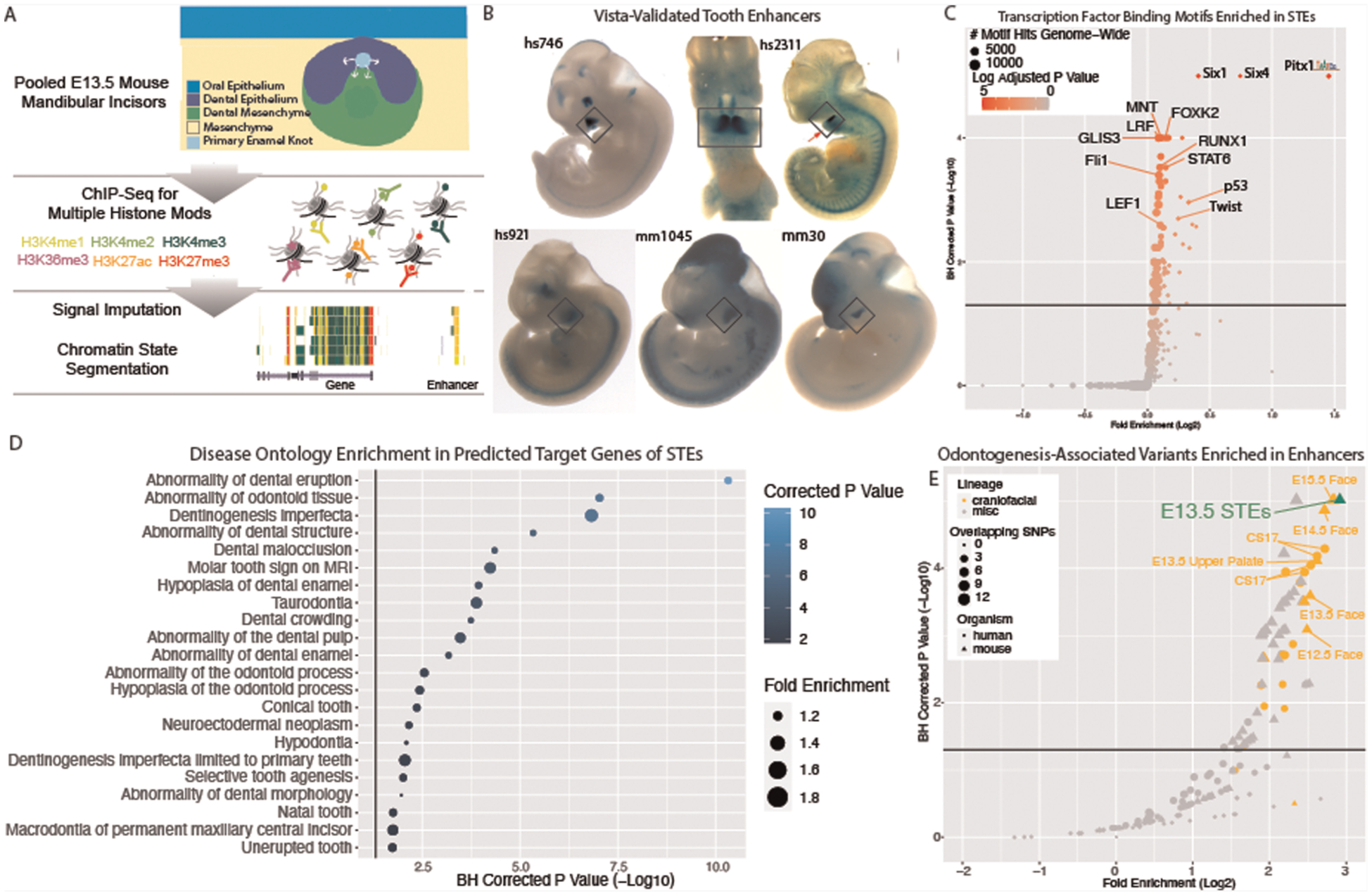
Enhancers of the developing tooth Are enriched for dental phenotype variants and target tooth development genes. (A) Schematic of ChIP-seq experiment and chromatin state annotation pipeline of E13.5 mouse mandibular incisors. (B) Selected VISTA-validated (58, 89) craniofacial enhancers which overlap TSEs. (C) Hypergeometric Optimization of Motif Enrichment (HOMER) (90) analysis of known transcription factor binding motifs enriched within tooth specific enhancers. Position weight matrices are demonstrated for each motif, obtained from JASPAR (133). (D) Dental disease ontology categories enriched in putative target genes of STEs, as assigned by Genomic Regions Enrichment of Annotations Tool (GREAT) (58). (E) GREGOR (60) analysis of enrichment of “Odontogenesis” (GO_0042476)-associated variants from GWAS Catalog in active enhancers annotated by 18 state chromatin segmentations genome-wide for samples from (40) (orange circles), Roadmap Epigenome (grey circles), and our own 18 state segmentations from ENCODE craniofacial ChIP-seq samples (orange triangles), E13.5 incisor (green) and other samples (grey triangles). Dotted line indicates log adjusted p-value threshold (1.3). Tooth specific enhancers (TSEs) do not pass significance threshold and cannot be evaluated for enrichment.
