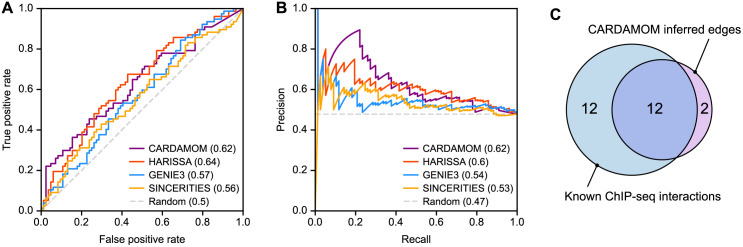
Fig 4. Comparison between inference methods and physical interactions derived from ChIP-seq.
The four directed GRN inference methods were applied to the experimental dataset from [22] restricted to a panel of 41 marker genes identified by the authors, and a reference network was obtained independently from edges supported by ChIP-seq data. As we only have access to physical interactions involving the retinoic acid (RA) stimulus or genes Pou5f1, Sox2, and Jarid2, the comparison only considers the 4 × 41 related edges. (A) Receiver operating characteristic (ROC) curve and corresponding area under the curve (AUROC) for each inference method. (B) Precision-recall (PR) curve and corresponding area under the curve (AUPR) for each inference method. (C) Venn diagram showing the overlap, for interactions involving the RA stimulus, between directed edges predicted by CARDAMOM and known physical interactions identified by ChIP-seq analysis.