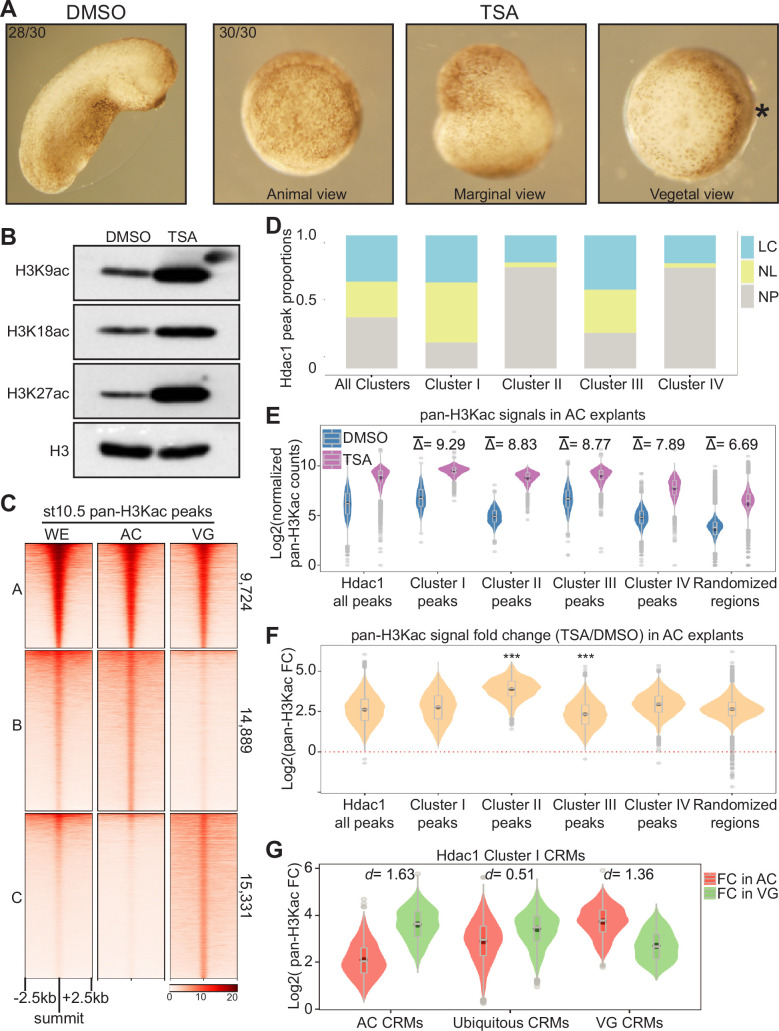
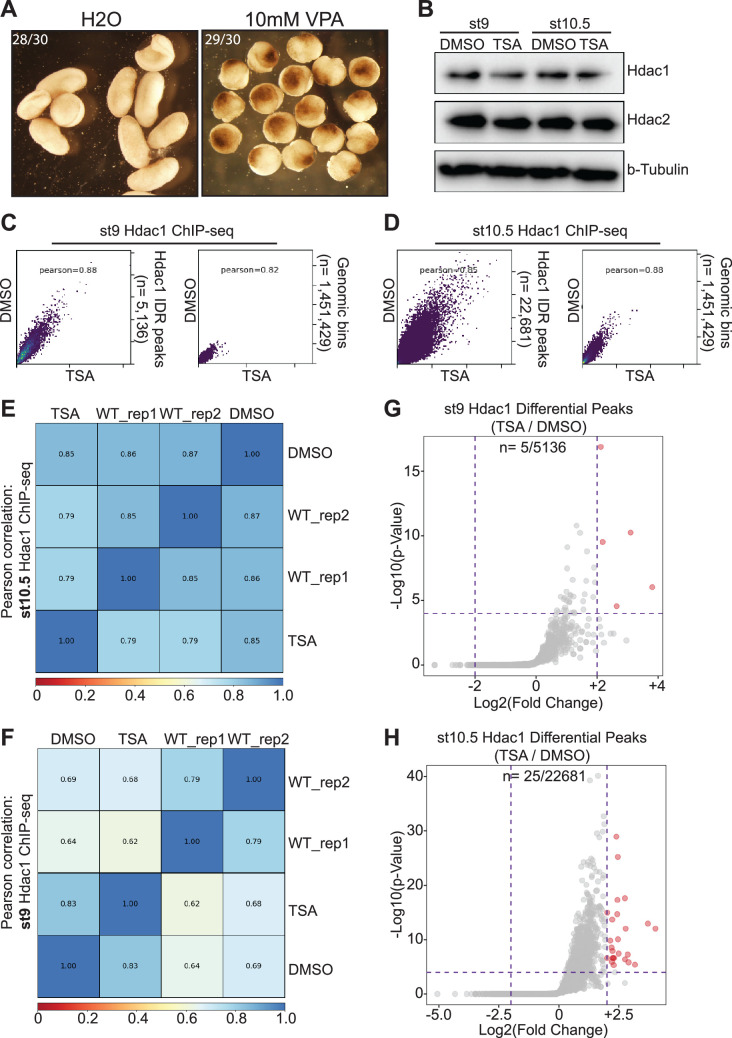
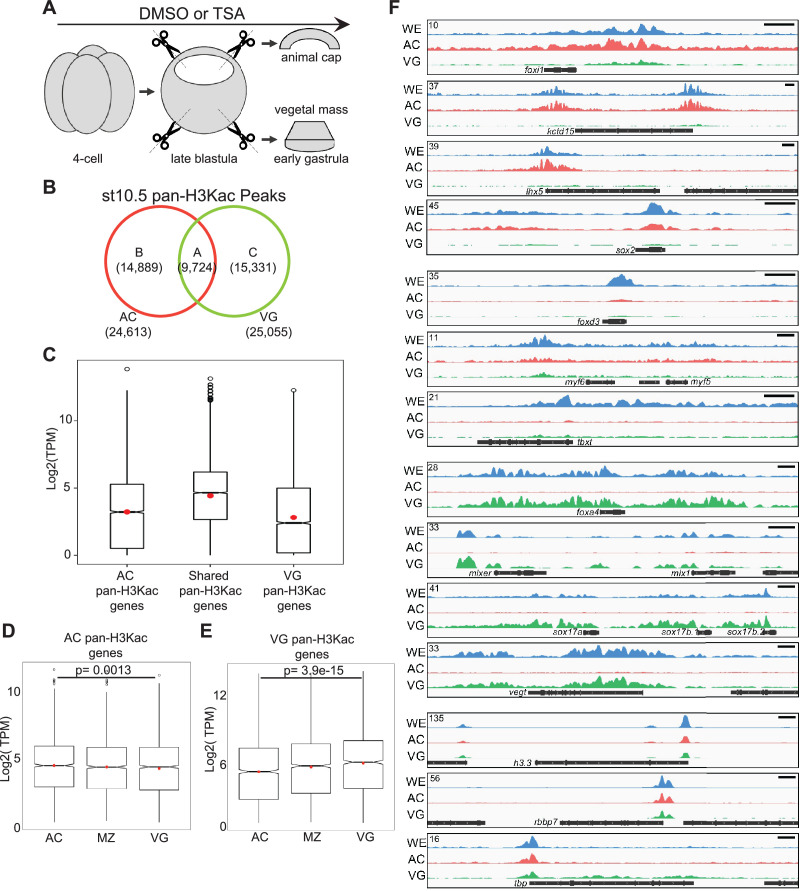
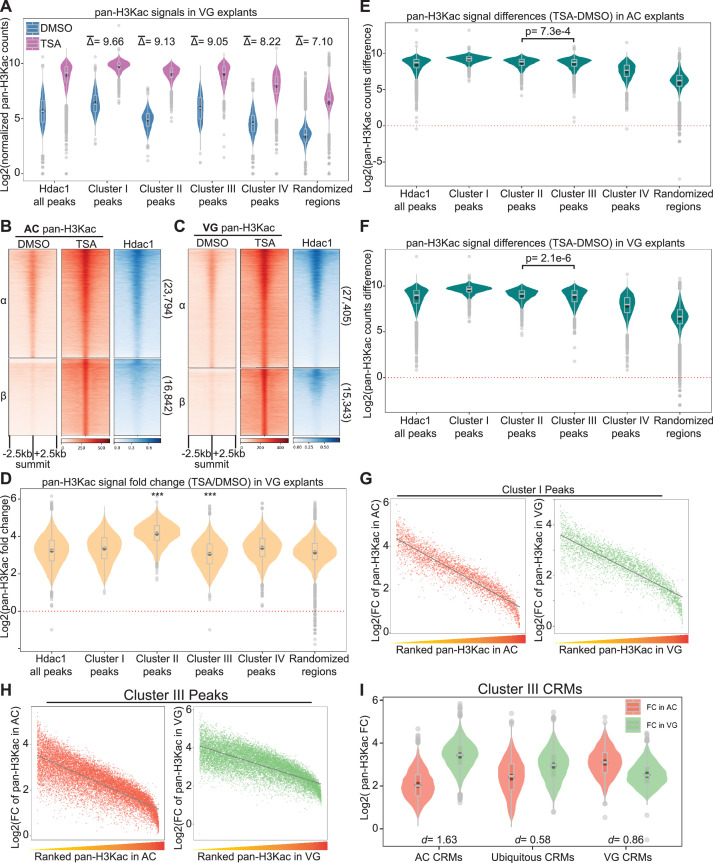
Figure 4. Hdac1 maintains differential H3 acetylomes between germ layers.
(A) Embryos treated with 100 nM Trichostatin A (TSA) displaying gastrulation defects 24 hr post-fertilization. Asterisk denotes the dorsal side containing the early blastopore lip. (B) Western blot analyses showing various histone acetylation modifications affected by HDAC inhibition. Anti-H3 is used as a loading control. (C) Clustered heatmap depicting signals of pan-H3Kac at st10.5 in whole embryos (WE), animal cap (AC), and vegetal mass (VG) cells. The signals are shown in a window of 5 kb centered on the summits of combined AC and VG peaks presented in descending order within each cluster. (D) Stacked bar graph representing proportions of localized (LC) versus non-localized (NL) pan-H3Kac signals found at Hdac1 cis-regulatory module (CRM) clusters in Figure 3C. A Hdac1 peak is considered to exhibit localized pan-H3Kac if it overlaps with either AC- or VG-specific pan-H3Kac peaks (Cluster B or C in C); a Hdac1 peak is considered to exhibit non-localized pan-H3Kac if it overlaps with pan-H3Kac peaks shared between AC and VG (Cluster A in C). NP: not overlap with any pan-H3Kac peak. (E) Spike-in normalized pan-H3Kac signals across Hdac1 CRM clusters (clusters in Figure 3C) in Dimethy Sulfoxide (DMSO)- or TSA-treated AC explants. represents the log2 scaled average differences of spike-in normalized pan-H3Kac signals between DMSO- and TSA-treated AC explants. Randomized genomic regions (n = 23,442) are used as the negative control. (F) Fold changes (FC) of pan-H3Kac signals at Hdac1 CRM clusters (clusters in Figure 3C) in DMSO- or TSA-treated AC explants. Red dotted line denotes the level of zero. *** denotes p < 0.001 (Student’s t-test). (G) Fold changes (FC) of pan-H3Kac signals in Cluster I of Hdac1 CRM clusters (clusters in Figure 3C) for each spatial CRM category. d denotes effect size calculated by Cohen’s d.