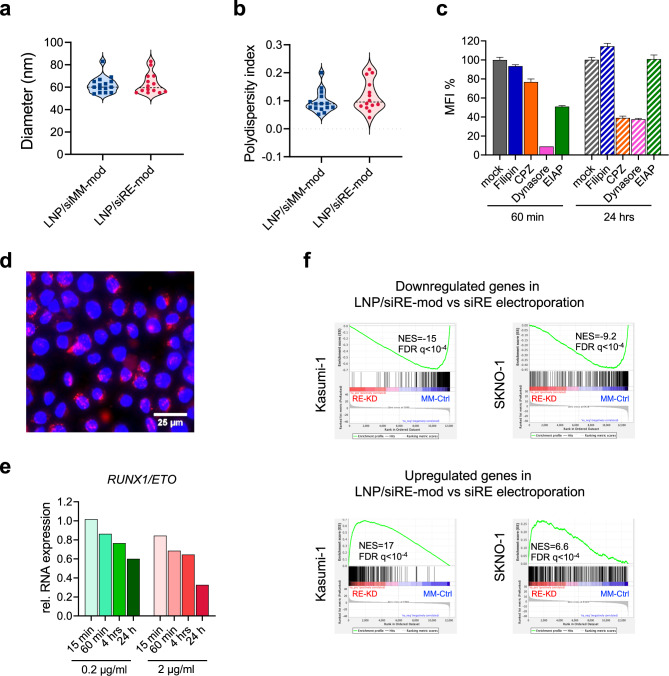
Fig. 2. Optimisation of lipid nanoparticle mediated RUNX1/ETO knockdown.
Measurement of LNPs/siRNAs diameter (a) and polydispersity (b) was performed after 1:100 dilution in PBS, each dot represents independent LNPs formulation. c, d Kasumi-1 cells were treated for 30 min with endocytosis inhibitors, then washed with PBS, resuspended in fresh medium, and exposed to Dil-labeled LNPs. The Dil fluorescence was measured by flow cytometry (c) 60 min and 24 h post-LNPs treatment (n = 4), and by fluorescence microscopy (d), the LNPs appear in red in the cytoplasm surrounding the DAPI-positive nucleus. e quantification of RUNX1/ETO expression in Kasumi-1 on day 3 relative to GAPDH (n = 1). Cells were treated with either 0.2 or 2 µg/ml LNPs/siRNAs for either 15 min, 60 min, 4 h or 24 h then cells were washed thrice with PBS to remove access LNPs. f gene enrichment analysis of Kasumi-1 and SKNO-1 following RUNX1/ETO knockdown by siRNA-LNPs treatment or electroporation (n = 3). Cells were either treated with 2 µg/ml siRNA-LNPs for 24 h then washed thrice with PBS and cultivated for further 2 days, or cells were electroporated with 200 nM siRNA. RNA-seq was performed on day 3.