Abstract
Viral variants of concern continue to arise for SARS-CoV-2, potentially impacting both methods for detection and mechanisms of action. Here, we investigate the effect of an evolving spike positive charge in SARS-CoV-2 variants and subsequent interactions with heparan sulfate and the angiotensin converting enzyme 2 (ACE2) in the glycocalyx. We show that the positively charged Omicron variant evolved enhanced binding rates to the negatively charged glycocalyx. Moreover, we discover that while the Omicron spike-ACE2 affinity is comparable to that of the Delta variant, the Omicron spike interactions with heparan sulfate are significantly enhanced, giving rise to a ternary complex of spike-heparan sulfate-ACE2 with a large proportion of double-bound and triple-bound ACE2. Our findings suggest that SARS-CoV-2 variants evolve to be more dependent on heparan sulfate in viral attachment and infection. This discovery enables us to engineer a second-generation lateral-flow test strip that harnesses both heparin and ACE2 to reliably detect all variants of concern, including Omicron.
Keywords: SARS-CoV-2, COVID-19, spike, heparin, heparan sulfate, ACE2, lateral-flow assay, biosensor, biomimetic, Brownian dynamics, electrostatic potential
Graphical abstract
Kim et al. uncover how SARS-CoV-2 variants evolve to be more dependent on heparan sulfate in viral attachment and infection. A combination of computational and experimental techniques reveal that increased positive charge on spike proteins leads to enhanced binding to heparan sulfate, informing the design of rapid diagnostics for COVID.
Introduction
Several SARS-CoV-2 variants of concern (VOCs) have emerged over the course of the COVID-19 pandemic, including Alpha, Beta, Gamma, Delta, and Omicron, the latter with its own sub-lineages BA.1, BA.2, BA.3, BA.4, and BA.5.1 , 2 , 3 Each of these VOCs are characterized by key mutations throughout the genome.1 , 2 , 3 The SARS-CoV-2 viral envelope is studded with approximately 30 homotrimer glycoproteins, called spike proteins, which play the primary role in initiating host-cell entry via their receptor binding domains (RBDs). Genomic mutations to the spike protein sequence have been implicated in increasing infectivity and/or immune escape.4 , 5 , 6 , 7 , 8 The Alpha, Beta, Delta, and Omicron BA.1 genomes, for example, contain 8, 8, 9, and 34 mutations in their spike mRNA sequences relative to the original “wild-type” (WT) 2019 strain9 , 10 , 11 , 12 , 13 (see Table S1 for complete list of mutations considered in this work).
The high number of sequence mutations characteristic of the Omicron variant presented a concern for potential impact on initial PCR detection.14 , 15 Rapid antigen detection was also impaired for Omicron variants even though these commercially available kits detect nucleocapsid proteins that incur a lower rate of mutation.14 , 15 Furthermore, the newer subvariants of Omicron, particularly BA.5, could completely escape from detection in current rapid kits.16 To overcome the reduced sensitivity of the rapid kits, the Food and Drug Administration recommended (August 11, 2022) repeated testing within 48 h.17 Rapid antigen detection of variant spike proteins is even more challenging considering the spike genome’s high mutation rate, often necessitating re-screening of spike antibodies, a step that can cause significant lag time behind the emergence of new variants.18 , 19 , 20
Elucidating the factors affecting spike binding kinetics and stability at the host-cell surface will help predict further mutations or gains of function, as well as aid in developing variant-specific antiviral therapies and better antigen testing platforms. As the SARS-CoV-2 virion approaches the cell surface (Step 1 in Scheme 1 A), it encounters the glycocalyx, a dense sugary matrix extending from the epithelial cell membrane.21 , 22 Heparan sulfate proteoglycans (HSPGs), key components of the glycocalyx, are known to serve as attachment factors for many viruses and likely to make first contact with SARS-CoV-2.23 , 24 , 25 HSPGs contain long, intrinsically disordered protein backbones decorated with longer (40–400 monomeric units) poly-sulfated and densely negatively charged glycosaminoglycans (GAGs).26 Heparan sulfate (HS) itself is biosynthesized natively in repeating dimeric units of N-acetyl-D-glucosamine and D-glucuronic acid; post-processing enzymes then add sulfate groups to certain positions along an HS sequence, and epimerization enzymes may convert some D-glucuronic acid monomers to L-iduronic acid. Neither sulfation nor epimerization reactions go to completion, thus there exist locally controlled regions of high/low sulfation/epimerization proportions further contributing to vast degree of glycocalyx heterogeneity. HS regions with particularly high proportions of sulfation and L-iduronic acid are referred to as “heparin-like” domains, calling on their similarity to short-chain medicinal heparin (HEP), which is almost completely sulfated and epimerized.26
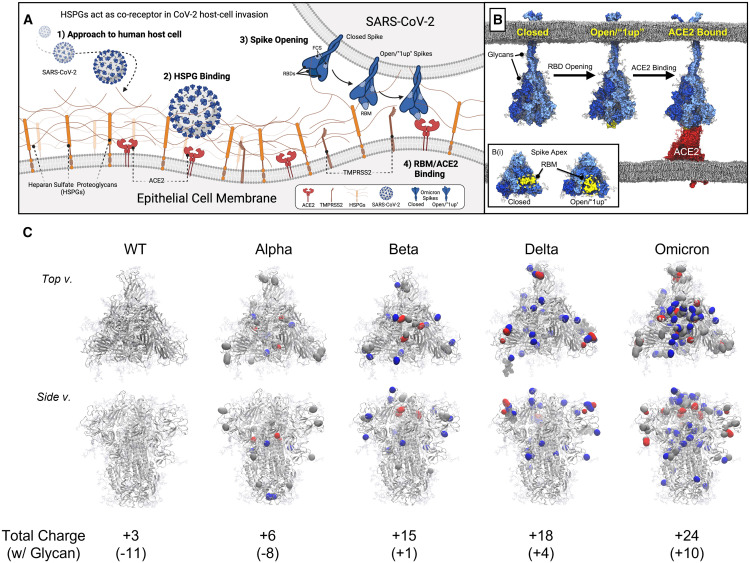
Scheme 1.
Illustration of SARS-CoV-2 host-cell invasion and mutations on the VOC spike proteins
(A) Scheme depicting initiation of host-cell invasion process through HS/ACE2/spike ternary complex formation via (1) SARS-CoV-2 approach to the human host-cell, (2) SARS-CoV-2 viral binding to heparan sulfate proteoglycans (HSPGs) on the host-cell membrane, (3) conformational change of SARS-CoV-2 spike proteins from a closed to open state, open spike demonstrating 1 receptor binding domain (RBD) in the “up” state with an exposed RBM, i.e., a “1up” spike, and finally (4) spike/ACE2 binding mediated by the receptor binding motif (RBM).
(B) Molecular models depicting steps 3 and 4 from (A).27 (B(i)) Inset illustrating an apical view of the spike head highlighting the RBM’s relative exposure in closed and open spike conformations.
(C) Mutations on the VOC spike protein, introduction of positive charges due to mutation on spike protein marked as blue, negative charges as red, and neutral as gray. Total charge of each spike protein head domain (residues 13 to 1,140, residues titrated to pH 7.4 with PROPKA28) given in parentheses next to the strain indicator.
Several studies in early 2020 first illustrated that the SARS-CoV-2 spike protein, particularly the RBD, can bind to HS and/or HEP,29 , 30 , 31 , 32 and that cellular HS was required for SARS-CoV-2 host-cell invasion.21 With these prior results, the second step in the viral invasion process comes into focus, as shown in Scheme 1A: Binding of the virion to HSPGs, likely through direct spike-HS interactions as shown by us33 and others.21 , 22 , 29 , 30 , 31 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 This step may increase virion residence time at the host-cell surface, thereby increasing the likelihood of encountering angiotensin converting enzyme 2 (ACE2), SARS-CoV-2’s primary host-cell receptor.38 , 39 , 43 The spike protein binds to ACE2 via a highly specialized interface within the RBD called the receptor binding motif (RBM).44 , 45 , 46 , 47 , 48 , 49 Each of the spike’s three RBDs (1 RBD per spike protomer, 3 RBDs per trimer spike complex, 1 RBM per RBD, 3 RBMs per trimer spike complex) may often occupy a “closed” or “down” conformational state wherein these RBMs are largely shielded from recognition.50 , 51 , 52 , 53 Thus, before spike-ACE2 binding can occur, at least one of these RBDs needs to emerge from its down/“shielded” state to an “up”/“open” state, to expose its RBM (Scheme 1B(i)).50 , 51 While this step (Scheme 1B) can occur anytime along the spike conformation switching, Clausen et al. report that short-chain HEP is capable of inducing spike RBDs to move into the up-state, suggesting that RBD opening could be induced after spike binding to HS and/or HEP within the glycocalyx.21 With the RBM exposed, ACE2 can bind, further stabilizing the virion at the host-cell surface. Finally, with the spike protein immobilized by HS and/or ACE2, TMPRSS2 can cleave the spike S2' site.54 , 55 , 56 , 57 , 58 Next, the S1 domain peels off from the S2, revealing the spike’s fusion peptide, which then penetrates the host-cell membrane and initiates membrane fusion.59 , 60 , 61
Recent studies have demonstrated that the Omicron SARS-CoV-2 virion relies less on membrane fusion as catalyzed by TMPRSS2 cleavage than earlier strains, and rather more on endocytosis. Indeed, syncytial formation—neighboring SARS-CoV-2 infected cells fusing together, a phenomenon indicative of TMPRSS2 activity—was reduced for Omicron-infected tissues.62 , 63 Furthermore, infection of TMPRSS2 knock-out cells by SARS-CoV-2 was increased for Omicron relative to other VOCs.63 , 64 This mechanistic change could be due to Omicron sequence mutations near the S2′ site causing decreased recognition by TMPRSS2.62 , 63 , 64 Furthermore, recent research suggests that mutations in the Delta and Omicron RBMs result in altered binding affinities to ACE2 compared with WT.65 , 66 , 67 , 68 , 69 , 70 Several past works have identified that charge-charge interactions heavily stabilize the WT spike-ACE2 interface,71 and that increasing spike charge over the course of SARS-CoV-2 evolution potentially increases electrostatic recognition of ACE2 at long range and increases immune escape,41 especially for Omicron, as its spike’s RBM is more positively charged than other spikes.42 While the role of evolving positive charge on spike proteins has begun to be unraveled for ACE2,41 , 42 , 65 , 66 , 67 , 68 , 69 , 70 this investigation was done in isolation from its required co-factor, HS.71 As HS is a long, negatively charged polysaccharide, the growing positive charge on spike virions, especially for Omicron, is expected to impact the stability of interactions with the HS-rich glycocalyx. In addition, since HS was previously shown to stabilize spike-ACE2 interactions, an altered affinity to HS may in turn also affect binding to ACE2.
Here, we probe the interactions between HS, ACE2, and the spike proteins of WT, Alpha, Beta, Delta, and Omicron SARS-CoV-2 variants. We propose how the positively charged Omicron spike may unlock a critical HS/ACE2 synergy. By harnessing the power of the primary and secondary cell receptors in the glycocalyx, we show how the performance of HEP-anchored test strips can co-evolve with the SARS-CoV-2 genome for robust and rapid sensing of Omicron. Finally, given that our synthetic glycocalyx test strips represent a minimal model for the cell surface, we discuss the potential implications our results may have on understanding SARS-CoV-2 infection dynamics at large.
Results and discussion
Spike mutations increase binding affinity to ACE2 and heparin
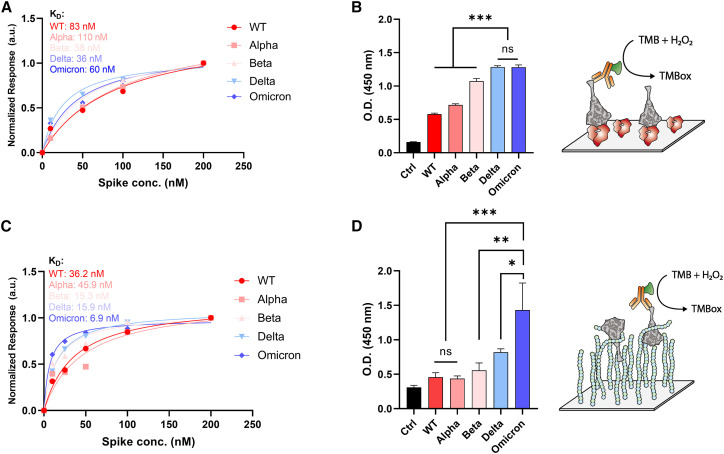
Previous work has demonstrated mixed results with respect to relative binding affinities between ACE2 and different spike protein sequences. Some groups report the Omicron spike protein binds to ACE2 with highest affinity, while others report there is no significant difference in binding affinity between all variants.65 , 66 , 67 , 68 , 69 , 70 Herein, we have used bio-layer interferometry (BLI), enzyme-linked immunosorbent assays (ELISA), and molecular dynamics (MD) simulations to estimate relative binding affinities between WT, Alpha, Beta, Delta, and Omicron BA.1 spike proteins to ACE2. BLI results indicate similar binding affinities within nanomolar range for all protein complexes, with Beta and Delta having the highest affinity, followed by Omicron BA.1, WT, and finally Alpha (Figures 1A and S1). ELISA results show a clearer trend: Delta and Omicron spikes similarly show higher affinity to ACE2 than all other spike proteins (Figure 1B). Our BLI and ELISA results were consistent with previous reports showing affinities of Delta and Omicron similarly increased compared with WT.65 , 66 , 69 , 70
Figure 1.
ACE2 and heparin binding affinity to VOC spike trimer
(A) Binding affinities of VOC spikes to dimeric ACE2 measured by BLI; KD values were calculated with steady-state analysis.
(B) Degree of bound complex formation for VOC spikes to dimeric ACE2 as measured by ELISA. TMB (3,3′,5,5′-tetramethylbenzidine) was used as a chromogen for ELISA.
(C) Binding affinities of VOC spikes to HEP measured by BLI; KD values calculated with steady-state analysis.
(D) Degree of bound complex formation for VOC spikes to HEP as measured by ELISA. Three independent tests were performed (n ≥ 3), and standard deviation from mean value was represented as an error bar in the graph. p values < 0.05 (∗), 0.01 (∗∗), and 0.001 (∗∗∗) were determined using a one-way ANOVA with Tukey’s post hoc test.
We also performed MD simulations of WT, Delta, and Omicron RBDs bound to ACE2 (supplemental experimental procedures). From these simulations we see interactions at the RBD/ACE2 interface that may explain increased binding affinity of Omicron spike relative to WT. Barros et al. discuss the relative contact frequencies of three subregions within the RBM to ACE2: the L3 loop, the central beta-strands, and the right-hand loops (Figure S2).47 They demonstrate that contacts formed between the central beta-strands and ACE2 are the strongest and maintain almost completely during their 3 replicas of 1 μs each of conformational sampling, whereas contacts formed between the L3 loop and right-hand loops to ACE2 are weaker, measured by decreased contacting frequency. From our MD simulations, we see the Omicron RBM (Figure S2), with 10 total mutations, both strengthens interactions to ACE2 within the central beta-strands (addition of salt bridge between Q493R to ACE2’s E35) but also increases stabilization within the weaker right-handed loops through an aromatic interaction (N501Y pi-stacking with ACE2’s Y41), and a tight hydrogen bonding network (Y505H tightly supporting a hydrogen bond between RBD’s Y495 and ACE2’s K353). These interactions, as also elucidated by Han et al.,72 potentially explain the strengthening of affinity between Omicron spike and ACE2 relative to the WT.
Next, we investigated the binding affinity between the variant spike proteins and HEP. BLI and ELISA results illustrate a significant increase in affinity between HEP and Omicron spike over Delta and other VOC spikes (Figures 1C, 1D, and S3). To probe differences in spike affinity to HEP at the molecular scale, we conducted extensive ensemble-based docking studies with AutoDock Vina73 , 74 to predict HEP binding modes to WT, Delta, and Omicron spikes in closed and open conformational states (supplemental experimental procedures). From ∼28,800 binding modes, we identified a total of 19 HEP binding “hotspots,” four of which were highly buried within the spike and 15 were located on the spike surface, thus accessible to long-chain HEP binding as would be seen within the glycocalyx (see Scheme S1, supplemental experimental procedures, and Figure S9 for complete descriptions of surface vs. buried sites). Relative affinities and populations of HEP binding modes at each of these sites were similar across the three spike variants (Figures S4–S11). In past work, we predicted three sites of high importance for interaction between the spike protein and HS: an RBD cleft site, an RBD patch site, and the furin cleavage site (FCS).33 Current ensemble-based docking simulations have confirmed the presence of these sites on WT and variant spikes and indicate there are no significant differences in binding affinities or number of binding modes in these sites between spike variants (Figures S4–S11 and supplemental experimental procedures). To determine the degree to which induced-fit effects within the RBD cleft, RBD patch, FCS, and potential binding at the RBM could impact affinity, we then conducted targeted flexible protein-flexible ligand docking studies with HEP and HS tetramers at each of these sites across WT, Delta, and Omicron variants with Schrödinger’s Induced Fit Docking protocol (Scheme S2 and supplemental experimental procedures).75 , 76 , 77 , 78 , 79 Again, there were no significant differences between average predicted binding energies for HEP or HS tetramers at each of these sites, across the three spike variants, even with global (MD enabled ensemble docking) and local (Schrödinger IFD) protein flexibility incorporated (Figure S12). This likely indicates that once an HS/HEP fragment finds a site on the spike surface, it is flexible enough to accommodate sequence mutations and maintain affinity at the surface. These docking results suggest that the increased binding affinity between HEP and SARS-CoV-2 Delta and Omicron spikes relative to WT, as observed with BLI and ELISA, most likely do not originate from site-specific changes.
Increased positive charge on Spike protein enhances rate of binding to ACE2 and Heparin
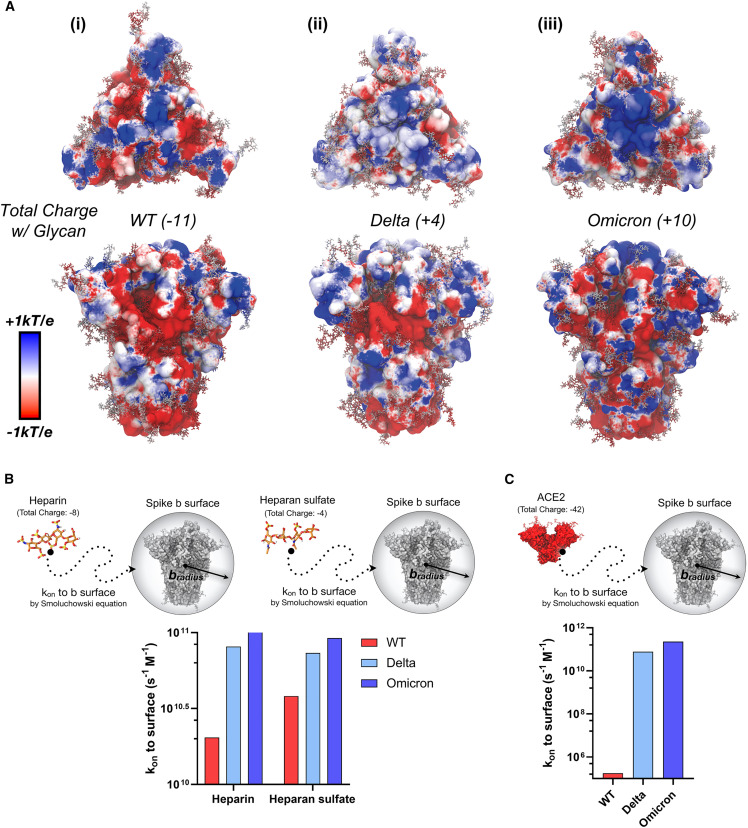
As previously noted by us43 , 80 and others,41 , 42 , 67 the spike protein is becoming more positively charged with each emerging VOC spike sequence (Scheme 1C). The total formal charge of the trimeric WT spike head domain (residues 13 to 1,140) at pH 7.4 is +3, Alpha is +6, Beta +15, Delta +18, and Omicron BA.1 +24. Several glycans on the spike ectodomain are also shown to be sialylated. Although a complete differentiation of glycan sialylation rate per spike sequence is far beyond the scope of this work, it is important to estimate the relative contribution of sialic acids to total spike head domain charge. Assuming a glycoprofile consistent with models from Casalino et al.50 (14 sialic acid residues on 57 head domain glycans) and described by Watanabe et al.,81 the total formal charge of the trimeric WT spike head domain with glycans is −11, Alpha is −8, Beta +1, Delta +4, and Omicron BA.1 +10 (Scheme 1C, Table S1). From this accounting of charge, it is evident that the spike protein head domains are increasing in total charge. In addition, to identify where positive and negative charges are most concentrated on the spike surface, we calculated dynamically averaged electrostatic potential maps of the WT, Delta, and Omicron (Figures 2A, S13, and supplemental experimental procedures). Interestingly, while the total spike charge is increasing, we also observed a clear redistribution of positive and negative charge across the spike surface between VOCs. Particularly, the Omicron spike surface exhibits a distinct redistribution of charges with a dramatic positively charged “bullseye” at its apex (Figure 2A(iii)). While the bullseye pattern is only clear in the closed state, the remapping of charge across the variant timeline is also demonstrated for 1up spike conformations (Figure S13). Increasing total charge of the spike protein, along with redistribution of charges on the spike surface have the potential to impact HS/HEP binding to spike within the glycocalyx.
Figure 2.
Dynamic electrostatic potential maps of spike trimer and Brownian dynamics simulation of HEP, HS, and ACE2 to spike trimer
(A) Dynamically averaged electrostatic potential maps collected from 50 ns of MD simulations for (i) WT, (ii) Delta, and (iii) Omicron spike proteins. The protein surfaces are colored according to average electrostatic potential at each site, ranging from −1 kBT/e (red) to +1 kBT/e (blue). Total charge of each spike protein head domain (residues 13 to 1,140, residues titrated to pH 7.4 with PROPKA28) given in parentheses next to the strain indicator; considering including a glycoprofile consistent with Casalino et al.50 and Watanabe et al.,81 WT, Delta, and Omicron spike proteins have total charges of −11, +4, and +10, respectively (14 sialic acids).
(B) Rate constant (kon) to b-surface calculated between heparin and heparan sulfate tetramer to WT, Delta, and Omicron spike proteins (each titrated to pH 7.4) with a corresponding scheme demonstrating system diffusion to the b-surface.
(C) Rate constant (kon) to b-surface calculated between and the ACE2 ectodomain (residues 18 to 734, pH 7.4) with a corresponding scheme demonstrating system diffusion to the b-surface. For (B) and (C), when calculating rate constants (kon) to the b-surface, receptor molecules are modeled as spheres defined by a b-radius, and total charge. The kon is calculated analytically according to the Smoluchowski equation (details in supplemental experimental procedures). Bars are colored according to a red to blue color scale normalized to VOC spike total charge with 14 sialylated glycans: WT (−11) in red, Delta (+4) in light blue, and Omicron (+10) in blue.58,82 It should be noted that error bars are not necessary for data presented in Figures 2B and 2C, as these values represent exact analytical solutions to the Smoluchowski equation.
To investigate the effects of spike total charge on HEP binding, we used Brownian dynamics (BD)83 , 82 simulations with Browndye83 , 82 to calculate rate constants (kon) to a “b-surface,” wherein the center of mass of a receptor molecule of interest defines the center of a sphere with a "b-radius” (Figure 2B). The receptor and ligand molecules, each containing partial atomic charges, approach one another from infinite space. In such a model, a kon between two molecules attaining an intermolecular distance less than the b-radius is largely driven by charge-charge interactions and can thus be solved numerically using the Smoluchowski equation.84 These results provide insight into long-range electrostatic interactions between molecules. We observe a dramatic increase in kon to the b-surface between an HEP tetramer (charge −8) and WT, Delta, and Omicron spike proteins, 2 × 1010 M−1s−1, 8 × 1010 M−1s−1, 1 × 1011 M−1s−1, respectively, (Figure 2B and supplemental experimental procedures). A similar trend is observed for the kon to the b-surface between a model HS tetramer (charge −4) and WT, Delta, and Omicron spikes (Figure 2B). In addition, seeing as the kon to the b-surface calculated for HS to WT was higher than that for HEP to WT, we predict that HS, owing to its decreased sulfation and charge, is likely to find and bind more quickly to WT spike surface than fully sulfated HEP domains. These results indicate that optimized long-range electrostatic interactions via spike mutations could dramatically impact the rate of SARS-CoV-2 viral approach to the glycocalyx (Step 1 in Scheme 1). Together with our docking results, which predicted very little difference between VOC spikes in HEP binding affinities at HEP binding hotspots, the BD results illustrate that increased affinities between HEP and Delta/Omicron spikes relative to WT, as seen by BLI and ELISA, may be due to kinetic selection allowing for increased encounters rather than site-specific differences in binding affinity. Furthermore, due to drastic difference in kon to b-surface for WT spike between HEP and HS tetrameric models (Figure 2B)—a trend that is nonexistent, if not reversed, for Delta and Omicron spike proteins—we predict that WT spike proteins have the potential to demonstrate increased selectivity for the less densely sulfated/charged heparin-like domains, while Delta and Omicron spike proteins would demonstrate little to no selectivity, or moderately increased selectivity for more densely sulfated/charged heparin-like domains.
Considering that ACE2’s dimeric ectodomain has a total charge at pH 7.4 of −42 (total formal charge with glycans at pH 7.4, −54)47 rates to ACE2, driven by long-range electrostatic interactions, may also be affected by increasing spike protein charge. Therefore, we also calculated rate constants (kons) to b-surfaces between the ACE2 ectodomain and WT, Delta, and Omicron spike proteins (supplemental experimental procedures). Interestingly, we see six orders of magnitude increase in kon between WT and Delta spikes to ACE2, followed by a one order of magnitude increase in kon between Delta and Omicron spikes to ACE2 (Figure 2C). As with the HS/HEP results, the increasing total charge of spike proteins may strengthen long-range electrostatic interactions to negatively charged ACE2. In addition, recall that binding affinities between SARS-CoV-2 VOC spikes and ACE2 are increasing (decreasing KD), but only moderately (Figures 1A and 1B). In sum, these BD results for HEP, HS, and ACE2 all point to kinetic fitness for interactions between the spike protein and negatively charged HEP/HS/ACE2 within the glycocalyx as a potential underlying evolutionary pressure driving SARS-CoV-2 spike sequence adaptation. Furthermore, the interactions of spike with other charged glycoprotein species and glycans within the glycocalyx such as neurophilin, CD147, GRP78, and sialic acids, could potentially be altered with the change in the charge on spike.
Remapping of positive charge distribution on Omicron surface maximizes heparin/ACE2 synergy
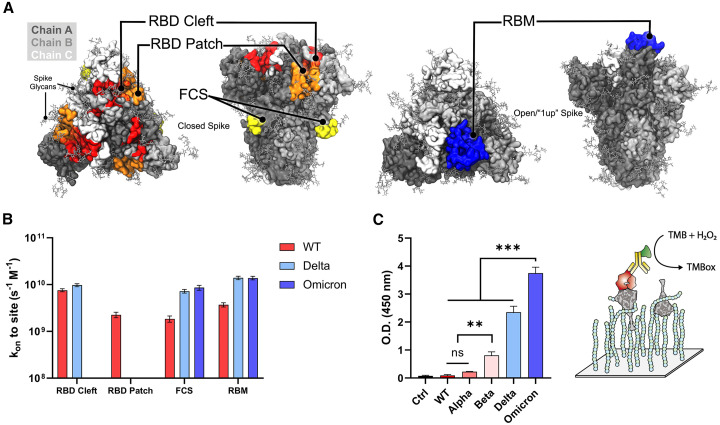
As can be seen from the dynamically averaged electrostatic potential maps in Figures 2A(i–iii) and S13, spike sequence mutations increase the total spike charge as well as redistribute surface patches of positive and negative charge. As a result, the site of first contact between HS/HEP and spike surface, i.e., a nucleation site for HS/HEP long-chain binding to the spike surface, could be altered on a per-VOC basis. To probe these changes, we again used BD simulations to investigate the rate of HS and HEP tetramer association, this time specifically to the RBM, RBD cleft, RBD patch, and the FCS sites (supplemental experimental procedures). We find that HEP tetramers associate differentially to spike surface sites due to mutations at each site (Figures 3A and 3B). Upon approaching a WT spike protein, our kinetic experiments indicate that HEP tetramers are most likely to associate with the RBD cleft site first, followed by the RBM, the RBD patch, and finally the FCS. However, upon approaching a Delta spike protein, HEP tetramers are most likely to find the RBM first, followed closely by the RBD cleft, and finally the FCS, with no observed transitions into the RBD patch. Similarly, when encountering Omicron spike proteins, HEP tetramers are most likely to first find and bind the RBM, followed by the FCS, with no observed transitions to the RBD cleft or patch sites. ESP maps of the Omicron RBM reveal that it is strongly positively charged, which likely supports the kinetic advantage for binding HEP at this site (Figure S13C(i)). These results indicate that redistribution of positive charges, especially for Omicron spikes, might cause a competition between HEP and ACE2 binding on the RBM site of the spike protein. However, at the cell surface, ternary complex formation among HS, spike, and ACE2 is potentially required for stabilization of the spike-ACE2 interface.21 Thus, we conducted ternary complex ELISA to identify whether HEP and ACE2 compete with one another for spike binding on a per-VOC basis (Figure 3C). While this ternary complex ELISA relies on both spike-HEP and spike-ACE2 binding, it could be hindered by decreased affinity at either interface. Strikingly, we observed a significant increase in the affinity of the Omicron spike-HEP-ACE2 ternary complex over all other variants, including Delta. It should be noted that when comparing Figure 3C with Figures 1B and 1D, the trend in ternary complex formation affinities compares more similarly to that seen in spike-HEP binary complex formation (Figure 1D) than spike-ACE2 complex formation (Figure 1B). Furthermore, the relatively similar affinities for Delta and Omicron spike proteins to ACE2 binding does not translate to a similar affinity in ternary complex formation for these two spike variants. These results indicate that remapping of positive charges, especially for Omicron, did not hinder the binding of HEP and ACE2 on spike protein. Instead, HEP and ACE2 can co-bind to the spike protein (Figure S13D shows the how 1up SARS-CoV-2 spike can accommodate ACE2 and HEP).
Figure 3.
Site-specific Brownian dynamics simulation and comparison of ternary complex formation between VOCs spike trimer
(A and B) Spike structures illustrating receptor binding domain (RBD) cleft, RBD patch, furin cleavage site (FCS), and the receptor binding motif (RBM) as designated sites targeted with BD simulations and (B) corresponding BD results shown as second-order rate constants between HEP and identified RBD cleft, RBD patch, FCS, and RBM sites.
(C) Degree of bound ternary complex formation for VOC spikes to HEP and ACE2 as measured by ELISA. Three independent tests were performed (n ≥ 3), and standard deviation from mean value is represented as an error bar in the graph. p values < 0.05 (∗), 0.01 (∗∗) and 0.001 (∗∗∗) were determined using a one-way ANOVA with Tukey’s post hoc test.
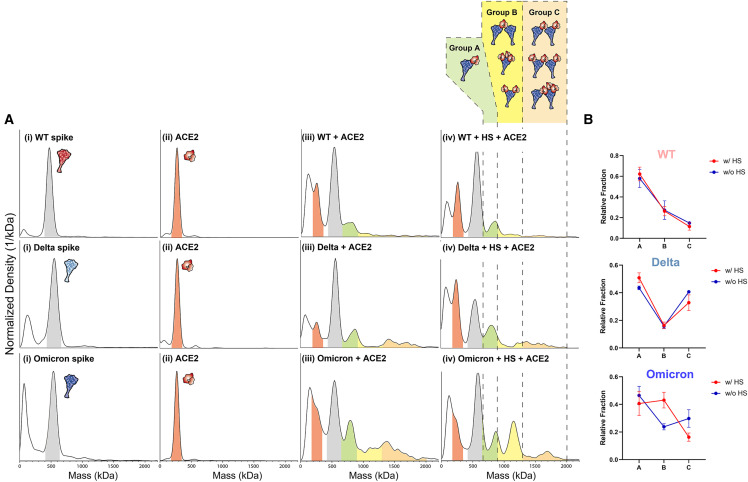
As HS has been reported to induce the open conformation of spike protein and enhance ACE2 binding, we further investigated the synergistic formation of ternary spike/HS/ACE2 complexes with mass photometry (MP). We then compared the effect of HS on formation of ternary complexes for WT, Delta, and Omicron spike proteins in the presence of dimeric ACE2, by measuring the mass distributions (Figures 4 and S14–S16 for repetition of MP results, supplemental experimental procedures).
Figure 4.
Synergistic formation of ternary spike/HS/ACE2 complexes visualized with mass photometry
(A) Mass photometry results comparing the WT, Delta, Omicron spikes binding to ACE2 and heparan sulfate (HS). (i) Mass distribution of WT, Delta, and Omicron spikes (mass range highlighted in gray), (ii) mass distribution of the dimeric ACE2 (mass range highlighted in red), (iii) mass distribution of spike protein + ACE2, and (iv) mass distribution of spike + HS + ACE2. To mimic the viral entry mechanism, HS was incubated with spike proteins first then followed by addition of ACE2. Possible ternary complexes are grouped in A (green), B (yellow), C (orange) based on their expected mass ranges. The molar ratio used in this study was spike:HS:ACE2 = 1:1:0.5.
(B) Fraction of ternary complex with or without HS for WT, Delta, and Omicron obtained by mass photometry. To calculate the fraction of these complexes, count numbers from each group (A, B, C) in Figure 4A were obtained. At least three independent experiments were performed, and error bars were calculated by the standard deviation of all experiments. Significance was calculated via multiple t test (unpaired) with Holm-Sidak method (α: 0.05) was performed.
The mass of each trimeric spike protein was measured to be around 560 kDa, and the mass of dimeric ACE2 was 240 kDa. Given that spike likely first encounters the extended tendrils of HS on approach to the human host-cell, we sequentially added first HS and then ACE2 to spike protein samples to mimic conditions at the cell surface. Co-incubation of spike with just ACE2 generated mass peaks for WT, Delta, and Omicron spike around 800 kDa, followed by signal density in higher mass ranges indicating that spike and ACE2 are interacting to form complexes at varying stoichiometric ratios (Figure 4A(iii)). Interestingly, while incubating spike and ACE2 with HS yielded very little differences in MP spectra for WT and Delta spikes compared with no-HS conditions, Omicron showed a significant increased population around 1,200 kDa under these testing conditions (Figure 4A (iv)), suggesting it plays a role in stabilizing a ternary spike-HS-ACE2. To assign possible stoichiometries to the emerging 1,200-kDa complex, we must understand the structural requirements for such assembly. Spike protein binding to ACE2 requires at least one spike RBD to be in the up conformation and a successful binding event between the two proteins is canonically considered as occurring between one ACE2 and one 1up spike. However, a spike protein with three RBDs in the up-state could accommodate binding of up to three ACE2 dimers and one ACE2 dimer could itself accommodate binding of up to two spike proteins.47 , 85 In addition, while dimeric ACE2 was used in this work, without the presence of B0AT-1—B0AT-1 is a sodium-dependent neutral amino acid transporter commonly found co-expressed and complexed with ACE2 in the gastrointestinal tract, B0AT-1 is often used to stabilize ACE2 during structural elucidation45 and its corresponding stabilization of the ACE2 interfacial neck domain—ACE2 in solution could exhibit more flexibility and adopt dual RBD binding modes as described by Xiao et al.86 While all such complexes are likely biologically relevant, the degree to which, and by what mechanism(s), spike and ACE2 form such complexes of “intermediate” stoichiometry is an open question. Thus, to parse our current mass photometry results, we have enumerated several configurations of spike-ACE2 complexes (illustration within Figure 4A) and divided such complexes into three groups based on their expected mass range: A (650–900 kDa), B (900–1,300 kDa), C (1,300–2,000 kDa). To compare the change in ternary complex distribution with or without HS, the fraction of each group (denoted as A, B, C) was calculated for each spike protein (Table S4–S6 for tables denoting fractions per group for Omicron, Delta, and WT spike proteins). As shown in Figure 4B, although the addition of HS slightly increased the population of group A type complexes for WT and Delta spikes. There was no significant change in degree of complex formation for type A along with type B and C for WT and Delta spikes. However, Omicron spikes showed significantly increased proportions of type B complexes in the presence of HS (Figure 4B). Considering that HS may stabilize spike RBDs in their up conformation, as reported by Clausen et al.,21 binding of multiple HS fragments to the Omicron spike could serve to recruit additional nearby ACE2s for binding, thereby increasing the population of group B type complexes (spike:ACE2 = 1:2, 1:3). In addition, an increased proportion of group B could also stem from one dimeric ACE2 binding two trimeric spike proteins, an interaction that could easily be facilitated by long chains of HS either binding one or both spike proteins. ACE2 bridging multiple spike proteins is likely an important factor governing complex formation at the cell surface, and HS has the potential to “hold” or cluster multiple spikes near ACE2 in preparation for multiplex binding.
To summarize all results presented thus-far: (1) binding affinity between SARS-CoV-2 VOC spikes to ACE2 are moderately increasing over the variant timeline, (2) site-specific affinities between SARS-CoV-2 VOC spikes to HEP dimers and tetramers have not changed significantly over the variant timeline, but (3) binding affinity between spike VOCs and long-chain HEP has increased over the variant timeline, (4) increasing total spike charge over the variant timeline may be increasing rates of HEP/HS/ACE2 to spike surfaces, (5) charge redistribution on the spike surface over the variant timeline may be altering HEP/HS nucleation sites in the context of long-chain binding interactions, and finally (6) Omicron has a particular ability to unlock a key HS/ACE2 synergy by increasing proportions of 1:2 and 1:3 spike:ACE2 complexes. At the cell surface, an individual spike glycoprotein will likely encounter both HS and ACE2. In what order, and by what mechanism(s) does the spike glycoprotein interact with and exploit the native functions of HS and ACE2 to enter the human host-cell, and how do mutations to the spike sequence affect this mechanism remain outstanding questions. As illustrated in Scheme 1, for a spike-ACE2 binding event to occur, the spike’s RBM needs to be sufficiently exposed, which only occurs when at least one of the spike protein’s RBDs moves from a “down”/“shielded” state to the “up”/“exposed” state. Clausen et al. report that short-chain HEP can increase proportions of ACE2s bound to the spike protein, suggesting HEP can facilitate RBD opening and ACE2 binding. Based on our results, we hypothesize that as the total formal charge of the spike protein increases so does the spike’s fitness for moving through and interacting with the negatively charged glycocalyx and ACE2 as shown in BLI, ELISA, and BD results. Moreover, as the spike approaches the glycocalyx, certain sites (i.e., the RBM and FCS) on VOC spikes may find and bind to HS more quickly than to others due to redistribution of charges on the spike surface. For example, in the case of the Omicron spike protein, given the rate constant for HEP binding is fastest to exposed RBMs, HS/HEP could increase the local concentration of 1up, 2up, and 3up Omicron spike proteins directly at the cell surface. While bound to the Omicron spike protein, HS would thereby stabilize spike in an attack-ready conformation while ACE2 arrives on the cell surface below. ACE2 could eventually displace HS from the RBM, which could in turn shift to one of the many other GAG-hotspots on the spike surface, including the FCS. In this fashion, the Omicron spike protein’s RBM could be capitalizing on HS/HEP’s capacity for kinetic selection, thereby increasing the localized concentration of ACE2-ready binding partners at the cell surface. Taken together, we suggest a mechanism by which SARS-CoV-2 variants evolve to better bind the co-receptor glycocalyx HS, which indirectly enhances its chances to bind and the stability of its interactions with the primary receptor, ACE2.
GlycoGrip test strips mimic host-cell surface to effectively detect evolved variants
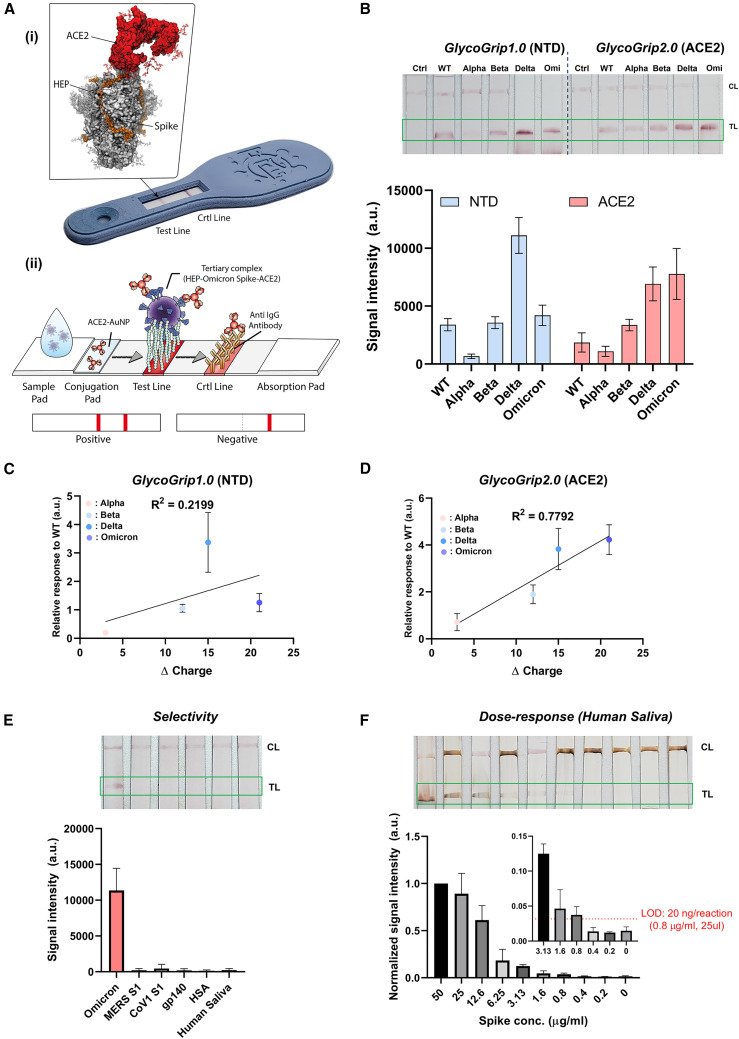
As discussed in the introduction, maintaining robust testing via rapid antigen and PCR detection platforms becomes a challenge during an actively progressing public health crisis such as the COVID-19 pandemic. Recently, we showed that the interaction between the host-cell surface glycopolymers and the spike glycoprotein can be exploited to detect SARS-CoV-2 in a rapid sandwich-style lateral-flow strip assay (LFSA).33 Our sensor termed GlycoGrip, was inspired by the interactions between SARS-CoV-2 virions and the glycocalyx. GlycoGrip uses long-chain heparin (HEP) to capture, and Au-nanoparticle conjugated anti-spike antibodies to signal for the presence of SARS-CoV-2 spike proteins. In our first generation of GlycoGrip, also known as GlycoGrip1.0, we used an N-terminal domain (NTD)-based anti-spike antibody to signal for the presence of WT, Alpha, Beta, and Delta spike proteins. In the current work, we leveraged our findings of a favorable tertiary complex formation between the Omicron spike protein, HS, and ACE2 to explore the potential of combining HEP as the capture agent, with ACE2 as the signaling probe: after all, if the virus utilizes these receptors to infect, GlycoGrip2.0 will leverage them to detect, (Figure 5 A (i,ii)).
Figure 5.
Analytical performance of GlycoGrip2.0
(A (i)) Image of a GlycoGrip prototype with callout image depicting a spike protein bound to both HEP (the GlycoGrip capture agent) and ACE2 (the signaling probe). (A (ii)) Schematic illustrating GlycoGrip’s capture and signaling of SARS-CoV-2 virions.
(B and C) Comparison of the GlycoGrip1.0 and GlycoGrip2.0 to VOCs with two different signaling probes (NTD Ab and ACE2; NTD Ab was previously used in GlycoGrip1.0), and (C) correlation between relative GlycoGrip1.0 signal intensity vs. change in spike charge; charge change calculated as VOC spike charge – WT spike charge, relative signal intensities plotted as ratio with respect to WT (same data as shown in B [NTD]).
(D) Correlation between relative GlycoGrip2.0 signal intensity vs. change in spike charge; charge change calculated as VOC spike charge—WT spike charge, relative signal intensities plotted as ratio with respect to WT (same data as shown in B [ACE2]).
(E) Selectivity of the GlycoGrip2.0 to relevant proteins including MERS, CoV1 spike, HIV envelope protein (gp140), human serum albumin (HSA), and human saliva.
(F) Dose-dependency results of Omicron detection using GlycoGrip2.0 with signal enhancement in human saliva condition. The limit of the detection was calculated by the blank + 3x (standard deviation of blank). At least three independent tests were performed (n ≥ 3) for GlycoGrip, and standard deviation from mean value was represented as an error bar in the graph.
For both generations of GlycoGrip, when a sample contains spike protein, a double-banded signal will appear on the lateral-flow strip: one band at the test line indicating ternary complex formation between HEP, spike, and the signaling probe, and one band at the control line indicating binary complex formation between the signaling probe and an anti-signaling probe antibody. We compared GlycoGrip1.0 and GlycoGrip2.0 against all VOC spike proteins (Figure 5B). We observed that, while GlycoGrip1.0 still signaled well for Omicron spike, signal intensity dropped significantly relative to Delta. However, the trend observed for WT, Alpha, Beta, and Delta spikes on GlycoGrip1.0 was similar to that reported previously.33 Notably, GlycoGrip2.0 elicited the strongest signal for Omicron spikes compared with GlycoGrip1.0. Moreover, we observed a clear trend: signal intensity on our GlycoGrip2.0 increases along with the variant timeline (Figure 5B) in a manner strikingly similar to the increase in total spike charge. Plotting this change in spike charge (i.e., total charge changes relative to WT spike) against relative signal intensity on our reconfigured GlycoGrip2.0, we see these two quantities correlate with one another: R 2 = 0.7792 (Figure 5E) while there was no clear trend for GlycoGrip1.0 (Figure 5C): R 2 = 0.2199. This correlation is striking given that, with HEP as the capture agent and ACE2 as the signaling probe, GlycoGrip2.0 can be seen as a simplified model for the cell-surface environment. These results indicate that, in contrast to antibody-based detection, our cell-surface mimetic sensor easily and effectively adapts to viral mutations, suggesting a novel paradigm shift in designing LFSA platforms to sensing viral antigens with high mutation rates.
Finally, we investigated the selectivity and sensitivity of our GlycoGrip2.0 specifically for detection of Omicron spike proteins. To determine the selectivity, we interrogated our sensor with related coronavirus (MERS, CoV1) and HIV (gp140) envelope proteins along with relevant complex proteins such as human serum albumin (HSA). To illustrate GlycoGrip’s feasibility when used against complex biologically relevant media, we also tested our sensor against a non-infected human saliva sample to check the false-positive signal from the complex biological samples. As shown in Figure 5E, GlycoGrip2.0 selectively captures and signals for Omicron spike proteins while not binding to related viral proteins, HSA, or other saliva matrix elements. Finally, Omicron was detectable as low as 40 ng/reaction (1.6 μg/mL, 25 μL, Figure S17) with ACE2, and 78 ng/reaction with NTD Abs. We then adopted a silver staining method to further enhance detection 4-fold for Omicron spikes: down to 10 ng/reaction (0.4 μg/mL, Figure S18). Last, we validated our sensor performance in human saliva samples. Human saliva contains various glycoproteins, including neutrophil elastase and histone H2A, which could interfere with the binding of either ACE2 or HEP to the spike protein.87 The limit of detection in saliva was estimated to be 20 ng/reaction (Figure 5F), which was comparable to detection in buffer conditions, demonstrating the power of our sensor to detect the virus in complex fluids (see Table S7 for comparison of GlycoGrip2.0 analytical performance with reported LFSA sensors). These results indicate GlycoGrip is selective for SARS-CoV-2 spike proteins, signals strongly in the presence of Omicron spikes, and is rapidly adaptable and deployable within the context of the ever-evolving COVID-19 public health crisis. Furthermore, as SARS-CoV-2 continues to adapt to niche evolutionary pressures within glycocalyx, GlycoGrip’s detection capacities will likely maintain and even strengthen without the need to change any of the sensor components.
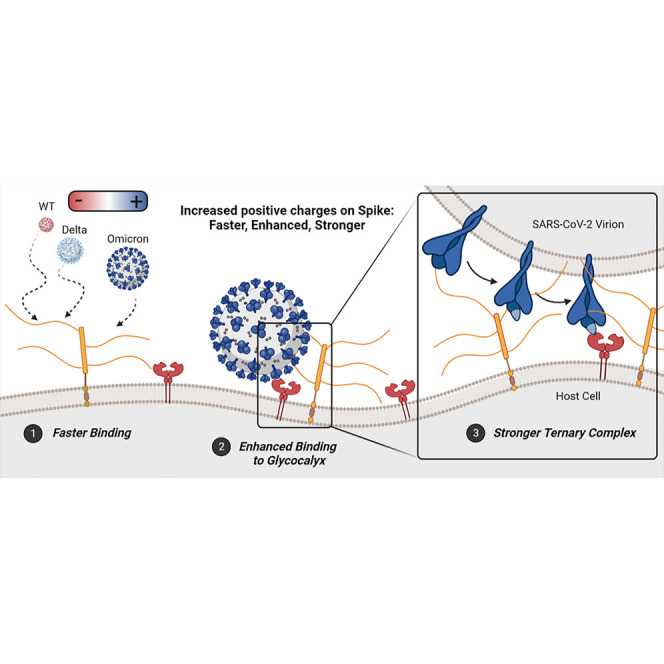
As the COVID-19 pandemic now progresses into its third year, public health experts continue to scan the epidemic-horizon for new variants. Delineating the environmental and immunological pressures driving SARS-CoV-2 genomic adaptation can help predict the likely range of future mutations, and the potential impacts of those mutations on infection, re-infection, hospitalization, and mortality rates. In this work, we revealed that increased total charge on the spike proteins of SARS-CoV-2 variants, due to the progressive addition of positively charged mutations, strengthens long-range electrostatic interactions with the negatively charged host-cell surface. Furthermore, we showed that the redistribution of positive and negative charges on evolving spike protein variants, particularly for Omicron, which adopts a striking “bullseye”-like patch of positive charge near the RBM, selectively enhances the rate and strength of HS binding to exposed RBMs. We thus hypothesize that Omicron SARS-CoV-2 kinetically increases the local concentration of ACE2-binding-ready spikes at the cell surface and unlocks a key synergy between HS and ACE2. We believe this remapping of positive charge on the SARS-CoV-2 spike protein is an evolutionary driver for the optimization of electrostatic interactions of spike proteins with both HS and ACE2, thereby increasing the rate of viral entry. With these conclusions in hand, one could predict that emerging variants will exhibit additional charge redistribution to further fine-tune these interactions and in turn increase SARS-CoV-2 infectivity.
Finally, we leveraged our findings of a favorable tertiary complex formation among the Omicron spike protein, HS, and ACE2 to develop the GlycoGrip2.0 sensor. We demonstrated GlycoGrip’s ability to “co-evolve” alongside the SARS-CoV-2 genome and we improved its detection of all VOCs (Video S1). By harnessing the primary (ACE2) and secondary (HS) cellular receptors in one sensor, GlycoGrip2.0 essentially serves as a minimal model of the glycocalyx environment, which may be a useful platform for viral surveillance. This highlights the advantage of glycocalyx-inspired sensing in a rapidly adapting public health crisis, as it is quickly reconfigurable and employable against evolving variants. As the COVID-19 pandemic is still ongoing, due to continuous evolution of the virus, glycocalyx-inspired LFSAs are likely to be a great benefit for global health monitoring power, not only for SARS-CoV-2 but for other rapidly mutating viral antigens.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ronit Freeman (ronifree@e-mail.unc.edu).
Materials availability
This study did not generate new unique materials.
Experimental methods
All methods including MD and BD simulations, docking studies, ESP calculations, BLI, and ELISA details can be found in the Supplemental Experimental Information.
Acknowledgments
The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper (http://www.tacc.utexas.edu). R.E.A. and R.F. acknowledge a Research Corporation for Science Advancement (COVID Initiative grant no. 27350) award. R.F. also acknowledges support from the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly, NSF RAPID (DMS-2028758), and the UNC Institute for Convergent Science Director’s Machete Award. R.E.A. acknowledges support from NIH GM132826, NSF RAPID MCB-2032054, a UC San Diego Moores Cancer Center 2020 SARS-COV-2 seed grant, and U19-AI171954 from NIAID. M.A.R. is supported by NIH T32 EB009380.
F.L.K., M.A.R., L.V., L.C., and R.E.A. acknowledge TACC Frontera and SDSC TSCC for their continued, unwavering support of our work over the past several years.
Author contributions
S.H.K., F.L.K., and M.A.R. contributed equally. S.H.K. and R.F. designed all experiments and conducted all assays. F.L.K., M.A.R., L.V., L.C., and R.E.A. designed all computational simulations and analyses. F.L.K., M.A.R., and L.V. conducted all computational simulations and analyses. L.C. contributed to the construction of the spike computational models. F.L.K. and M.A.R. constructed and designed all computational model images. S.H.K., F.L.K., M.A.R., R.E.A., and R.F. wrote the manuscript with input from all the authors. M.J.P. provided lung biology insights and edited the paper. R.E.A. and R.F. provided foundational guidance on experimental design and analyses.
Declaration of interests
The authors declare the following patent: Glycosaminoglycan articles and methods relating thereof. Inventors: S.H.K, R.F. Application number PCT/US22/79641 was submitted on this work.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrp.2023.101346.
Supplemental information
Data and code availability
The authors declare that all data supporting the findings of this study are available within this article and its supplemental information files. Any additional information reported in this paper is available from the lead contact on request. All structures relating to simulations described herein will also be made freely available to download on the AmaroLab Web site. The following files are available at https://amarolab.ucsd.edu/covid19.php: structures (psf/pdb formats) of all docking results described herein; APBS input files for all computational calculations done herein including docking and electrostatic potential and binding energy calculations; BD input files for all Brownian dynamics simulations conducted herein. Any questions or additional information needed to access these files will be handled by the lead contact on request.
References
- 1.World Health Organization . WHO Coronavirus (COVID-19) Dashboard. 2020. [Google Scholar]
- 2.World Health Organization . WHO activity; 2021. Tracking SARS-CoV-2 Variants of Concern. [Google Scholar]
- 3.Center for Disease Control and Prevention . CDC Variants and Surveillance; 2022. SARS-CoV-2 Variant Classifications and Definitions. [Google Scholar]
- 4.Bashor L., Gagne R.B., Bosco-Lauth A.M., Bowen R.A., Stenglein M., VandeWoude S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2105253118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evolutionary insight into the emergence of SARS-CoV-2 variants of concern. Nat. Med. 2022;28:1357–1358. doi: 10.1038/s41591-022-01892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Van Domselaar G., Wu J., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maher M.C., Bartha I., Weaver S., di Iulio J., Ferri E., Soriaga L., Lempp F.A., Hie B.L., Bryson B., Berger B., et al. Predicting the mutational drivers of future SARS-CoV-2 variants of concern. Sci. Transl. Med. 2022;14:eabk3445. doi: 10.1126/scitranslmed.abk3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., et al. COVID-19 Genomics UK COG-UK Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., De Marco A., Franko N., Tilles S.W., Logue J., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- 10.Public Health England . Technical Briefing; 2020. Public Health England Investigation of Novel SARS-COV-2 Variant 202012/01. [Google Scholar]
- 11.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.-K., Adams-Quack P., Bacher M., Finlayson A., et al. 1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha L.B., Foster C., Rawlinson W., Tedla N., Bull R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: implications for immune escape and transmission. Rev. Med. Virol. 2022;32 doi: 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohapatra R.K., Kandi V., Verma S., Dhama K. Challenges of the omicron (B.1.1.529) variant and its lineages: a global perspective. Chembiochem. 2022;23 doi: 10.1002/cbic.202200059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao W., Zhang W., Fang X., Yu D., Wang X. Challenges of SARS-CoV-2 omicron variant and appropriate countermeasures. J. Microbiol. Immunol. Infect. 2022;55:387–394. doi: 10.1016/j.jmii.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuzinger K., Roloff T., Egli A., Hirsch H.H. Impact of SARS-CoV-2 omicron on rapid antigen testing developed for early-pandemic SARS-CoV-2 variants. Microbiol. Spectr. 2022;10:e0200622–e0202022. doi: 10.1128/spectrum.02006-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.United States Food and Drug Administration . FDA Safety Communication; 2022. At-Home COVID-19 Antigen Tests-Take Steps to Reduce Your Risk of False Negative. [Google Scholar]
- 18.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 21.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue J., Jin W., Yang H., Faulkner J., Song X., Qiu H., Teng M., Azadi P., Zhang F., Linhardt R.J., Wang L. Heparan sulfate facilitates spike protein-mediated SARS-CoV-2 host cell invasion and contributes to increased infection of SARS-CoV-2 G614 mutant and in lung cancer. Front. Mol. Biosci. 2021;8:649575. doi: 10.3389/fmolb.2021.649575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cagno V., Tseligka E.D., Jones S.T., Tapparel C. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11 doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stencel-Baerenwald J.E., Reiss K., Reiter D.M., Stehle T., Dermody T.S. The sweet spot: defining virus–sialic acid interactions. Nat. Rev. Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D., Esko J.D. Demystifying heparan sulfate–protein interactions. Annu. Rev. Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casalino L., Dommer A.C., Gaieb Z., Barros E.P., Sztain T., Ahn S.-H., Trifan A., Brace A., Bogetti A.T., Clyde A., et al. AI-driven multiscale simulations illuminate mechanisms of SARS-CoV-2 spike dynamics. The International Journal of High Performance Computing Applications. 2021;10943420211006452 doi: 10.1177/10943420211006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsson M.H.M., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. Journal of Chemical Theory and Computation. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 29.Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Gandhi N.S., Guimond S.E., Miller G.J., Meneghetti M.C.Z., Nader H.B., et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb. Haemost. 2020;120:1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Chopra P., Li X., Bouwman K.M., Tompkins S.M., Wolfert M.A., de Vries R.P., Boons G.-J. Heparan sulfate proteoglycans as attachment factor for SARS-CoV-2. ACS Cent. Sci. 2021;7:1009–1018. doi: 10.1021/acscentsci.1c00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., Fu L., Dordick J.S., Woods R.J., Zhang F., Linhardt R.J. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res. 2020;181 doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalra R.S., Kandimalla R. Engaging the spikes: heparan sulfate facilitates SARS-CoV-2 spike protein binding to ACE2 and potentiates viral infection. Signal Transduct. Target. Ther. 2021;6:39. doi: 10.1038/s41392-021-00470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S.H., Kearns F.L., Rosenfeld M.A., Casalino L., Papanikolas M.J., Simmerling C., Amaro R.E., Freeman R. GlycoGrip: cell surface-inspired universal sensor for betacoronaviruses. ACS Cent. Sci. 2022;8:22–42. doi: 10.1021/acscentsci.1c01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuurs Z.P., Hammond E., Elli S., Rudd T.R., Mycroft-West C.J., Lima M.A., Skidmore M.A., Karlsson R., Chen Y.-H., Bagdonaite I., et al. Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Comput. Struct. Biotechnol. J. 2021;19:2806–2818. doi: 10.1016/j.csbj.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., McCandless M.G., Jin W., Liu H., Sharma P., Linhardt R.J. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J. Virol. 2021;95:e01987-20–e01920. doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Chen C.Z., Swaroop M., Xu M., Wang L., Lee J., Wang A.Q., Pradhan M., Hagen N., Chen L., et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;6:80. doi: 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paiardi G., Richter S., Oreste P., Urbinati C., Rusnati M., Wade R.C. The binding of heparin to spike glycoprotein inhibits SARS-CoV-2 infection by three mechanisms. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A., Berniak K., Wojarski J., Zeglen S., Baster Z., et al. Entry of human coronavirus NL63 into the cell. J. Virol. 2018;92:e01933-17–e01917. doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascarella S., Ciccozzi M., Bianchi M., Benvenuto D., Cauda R., Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: a hint to higher transmissibility? J. Med. Virol. 2022;94:1277–1280. doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 41.Gan H.H., Zinno J., Piano F., Gunsalus K.C. Omicron spike protein has a positive electrostatic surface that promotes ACE2 recognition and antibody escape. Front. Virol. 2022;2 [Google Scholar]
- 42.Nie C., Sahoo A.K., Netz R.R., Herrmann A., Ballauff M., Haag R. Charge matters: mutations in omicron variant favor binding to cells. Chembiochem. 2022;23 doi: 10.1002/cbic.202100681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearns F.L., Sandoval D.R., Casalino L., Clausen T.M., Rosenfeld M.A., Spliid C.B., Amaro R.E., Esko J.D. Spike-heparan sulfate interactions in SARS-CoV-2 infection. Curr. Opin. Struct. Biol. 2022;76 doi: 10.1016/j.sbi.2022.102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 45.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barros E.P., Casalino L., Gaieb Z., Dommer A.C., Wang Y., Fallon L., Raguette L., Belfon K., Simmerling C., Amaro R.E. The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys. J. 2021;120:1072–1084. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sztain T., Ahn S.-H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahbar Saadat Y., Hosseiniyan Khatibi S.M., Zununi Vahed S., Ardalan M. Host serine proteases: a potential targeted therapy for COVID-19 and influenza. Front. Mol. Biosci. 2021;8:725528. doi: 10.3389/fmolb.2021.725528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., Hall W.W., Orba Y., Sawa H. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papa G., Mallery D.L., Albecka A., Welch L.G., Cattin-Ortolá J., Luptak J., Paul D., McMahon H.T., Goodfellow I.G., Carter A., et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Essalmani R., Jain J., Susan-Resiga D., Andréo U., Evagelidis A., Derbali R.M., Huynh D.N., Dallaire F., Laporte M., Delpal A., et al. Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. J. Virol. 2022;96:00128222–e100122. doi: 10.1128/jvi.00128-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koppisetti R.K., Fulcher Y.G., Van Doren S.R. Fusion peptide of SARS-CoV-2 spike rearranges into a wedge inserted in bilayered micelles. J. Am. Chem. Soc. 2021;143:13205–13211. doi: 10.1021/jacs.1c05435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., Cantoni D., Scott S., Logan N., Ashraf S., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peacock T.P., Brown J.C., Zhou J., Thakur N., Sukhova K., Newman J., Kugathasan R., Yan A.W.C., Furnon W., De Lorenzo G., et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. Preprint at. [DOI] [Google Scholar]
- 65.Hoffmann M., Zhang L., Pöhlmann S. Omicron: master of immune evasion maintains robust ACE2 binding. Signal Transduct. Target. Ther. 2022;7:118. doi: 10.1038/s41392-022-00965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Socher E., Heger L., Paulsen F., Zunke F., Arnold P. Molecular dynamics simulations of the delta and omicron SARS-CoV-2 spike – ACE2 complexes reveal distinct changes between both variants. Comput. Struct. Biotechnol. J. 2022;20:1168–1176. doi: 10.1016/j.csbj.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotten M., Phan M.V.T. Evolution to increased positive charge on the viral spike protein may be part of the adaptation of SARS-CoV-2 to human transmission. bioRxiv. 2022 doi: 10.1101/2022.07.30.502143. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta P., Ravi V., Devi P., Maurya R., Parveen S., Mishra P., Yadav A., Swaminathan A., Saifi S., Khare K., et al. Mutational dynamics across VOCs in International travellers and Community transmission underscores importance of Spike-ACE2 interaction. Microbiol. Res. 2022;262 doi: 10.1016/j.micres.2022.127099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Costa C.H.S., de Freitas C.A.B., Alves C.N., Lameira J. Assessment of mutations on RBD in the spike protein of SARS-CoV-2 Alpha, delta and omicron variants. Sci. Rep. 2022;12:8540. doi: 10.1038/s41598-022-12479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S., Liu Y., Ziarnik M., Seo S., Cao Y., Zhang X.F., Im W. Binding of human ACE2 and RBD of Omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern. J. Comput. Chem. 2023;44:594–601. doi: 10.1002/jcc.27025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X., Bie L., Gao J. Structural insights into the cofactor role of heparin/heparan sulfate in binding between the SARS-CoV-2 spike protein and host angiotensin-converting enzyme II. J. Chem. Inf. Model. 2022;62:656–667. doi: 10.1021/acs.jcim.1c01484. [DOI] [PubMed] [Google Scholar]
- 72.Han P., Li L., Liu S., Wang Q., Zhang D., Xu Z., Han P., Li X., Peng Q., Su C., et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022;185:630–640.e10. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schrodinger Release 2022-3 . Schrödinger, LLC; 2022. Induced Fit Docking Protocol. [Google Scholar]
- 76.Sherman W., Beard H.S., Farid R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006;67:83–84. doi: 10.1111/j.1747-0285.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 77.Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 78.Farid R., Day T., Friesner R.A., Pearlstein R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg. Med. Chem. 2006;14:3160–3173. doi: 10.1016/j.bmc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 79.Schrödinger Release 2021-3. Glide . Schrödinger, LLC; 2021. [Google Scholar]
- 80.Carl Zimmer J.C. The New York times. The Coronavirus in a Tiny Drop. 2021 [Google Scholar]
- 81.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huber G.A., McCammon J.A. Browndye: a software package for Brownian dynamics. Comput. Phys. Commun. 2010;181:1896–1905. doi: 10.1016/j.cpc.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huber G.A., McCammon J.A. Brownian dynamics simulations of biological molecules. Trends Chem. 2019;1:727–738. doi: 10.1016/j.trechm.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chavanis P.-H. The generalized stochastic Smoluchowski equation. Entropy. 2019;21:1006. doi: 10.3390/e21101006. [DOI] [Google Scholar]
- 85.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao T., Lu J., Zhang J., Johnson R.I., McKay L.G.A., Storm N., Lavine C.L., Peng H., Cai Y., Rits-Volloch S., et al. A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat. Struct. Mol. Biol. 2021;28:202–209. doi: 10.1038/s41594-020-00549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshizato K., Taira T., Sato-Matsubara M., Sekiguchi S., Yabunaka Y., Kira Y., Ohashi T., Daikoku A., Ofusa K., Kadono C., et al. Cloaking the ACE2 receptor with salivary cationic proteins inhibits SARS-CoV-2 entry. J. Biochem. 2022;172:205–216. doi: 10.1093/jb/mvac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within this article and its supplemental information files. Any additional information reported in this paper is available from the lead contact on request. All structures relating to simulations described herein will also be made freely available to download on the AmaroLab Web site. The following files are available at https://amarolab.ucsd.edu/covid19.php: structures (psf/pdb formats) of all docking results described herein; APBS input files for all computational calculations done herein including docking and electrostatic potential and binding energy calculations; BD input files for all Brownian dynamics simulations conducted herein. Any questions or additional information needed to access these files will be handled by the lead contact on request.