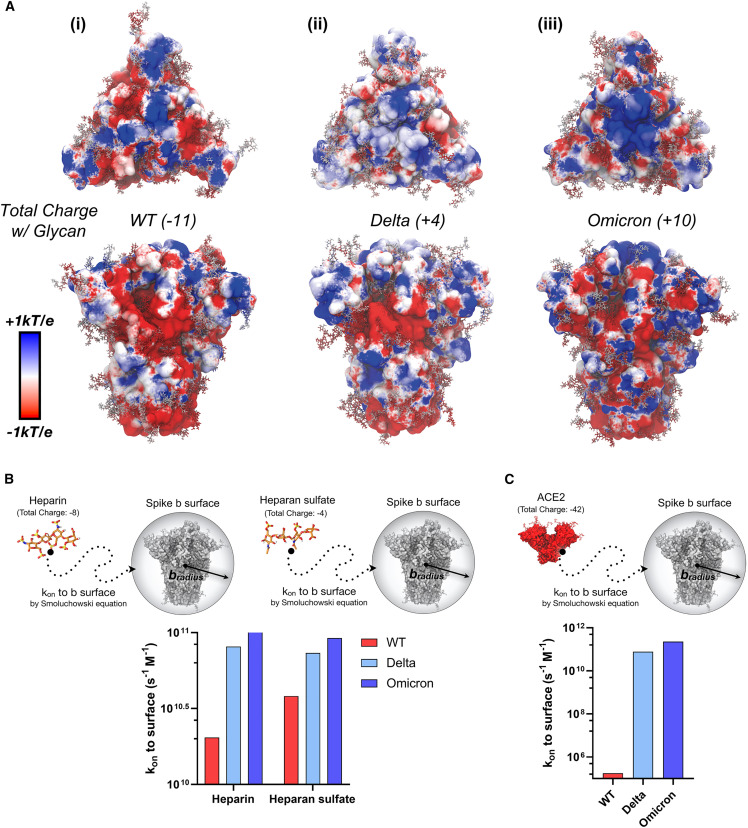
Figure 2.
Dynamic electrostatic potential maps of spike trimer and Brownian dynamics simulation of HEP, HS, and ACE2 to spike trimer
(A) Dynamically averaged electrostatic potential maps collected from 50 ns of MD simulations for (i) WT, (ii) Delta, and (iii) Omicron spike proteins. The protein surfaces are colored according to average electrostatic potential at each site, ranging from −1 kBT/e (red) to +1 kBT/e (blue). Total charge of each spike protein head domain (residues 13 to 1,140, residues titrated to pH 7.4 with PROPKA28) given in parentheses next to the strain indicator; considering including a glycoprofile consistent with Casalino et al.50 and Watanabe et al.,81 WT, Delta, and Omicron spike proteins have total charges of −11, +4, and +10, respectively (14 sialic acids).
(B) Rate constant (kon) to b-surface calculated between heparin and heparan sulfate tetramer to WT, Delta, and Omicron spike proteins (each titrated to pH 7.4) with a corresponding scheme demonstrating system diffusion to the b-surface.
(C) Rate constant (kon) to b-surface calculated between and the ACE2 ectodomain (residues 18 to 734, pH 7.4) with a corresponding scheme demonstrating system diffusion to the b-surface. For (B) and (C), when calculating rate constants (kon) to the b-surface, receptor molecules are modeled as spheres defined by a b-radius, and total charge. The kon is calculated analytically according to the Smoluchowski equation (details in supplemental experimental procedures). Bars are colored according to a red to blue color scale normalized to VOC spike total charge with 14 sialylated glycans: WT (−11) in red, Delta (+4) in light blue, and Omicron (+10) in blue.58,82 It should be noted that error bars are not necessary for data presented in Figures 2B and 2C, as these values represent exact analytical solutions to the Smoluchowski equation.