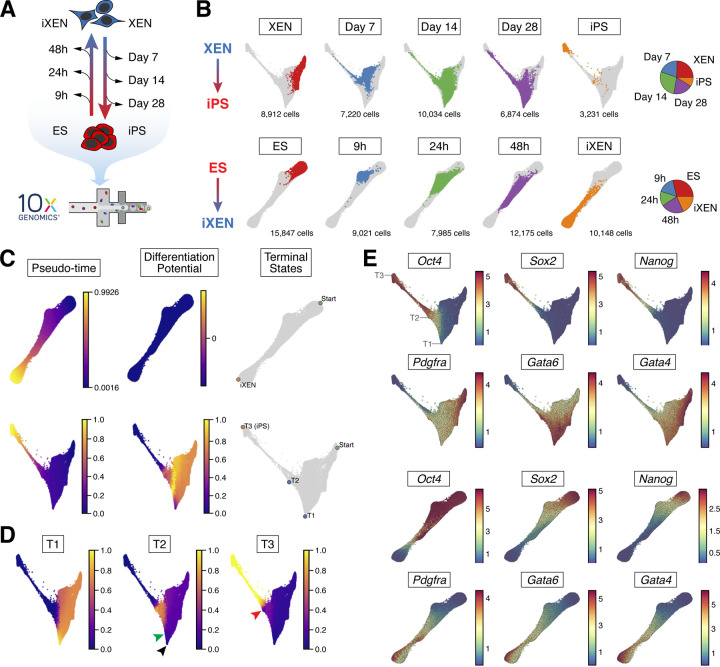
Figure 3. scRNA-seq analyses of XEN-to-iPS and ES-to-iXEN conversions.
(A) Experimental scheme outlining the cells and lineage conversion timepoints assayed by scRNA-seq.
(B) Force-directed layouts showing pooled timepoints of XEN-to-iPS (top) and ES-to-iXEN (bottom) conversion in a single trajectory. Individual plots highlight the distribution of single timepoints across the trajectory. (Right) Pie chart representations of the proportion of cells representing each collection timepoint relative to the entire cohort per conversion trajectory.
(C) Palantir determined pseudotime ordering, terminal states, and differentiation potential of ES-to-iXEN (top) and XEN-to-iPS (bottom) trajectories.
(D) Branch probabilities of terminal states determined by Palantir in the XEN-to-iPS trajectory. Black arrowhead indicates cells at T1 with low probability of differentiating to T2. Green arrowhead indicates where T2 probability increases, coinciding with Oct4 expression. Red arrowhead indicates cells at T2, where they have a non-zero probability of acquiring the T3 state.
(E) Gene expression patterns of XEN and pluripotency-associated markers. Each cell is colored on the basis of its MAGIC imputed expression level for the indicated gene. Locations of T1, T2 and T3 terminal states are indicated for the XEN-to-iPS trajectory.