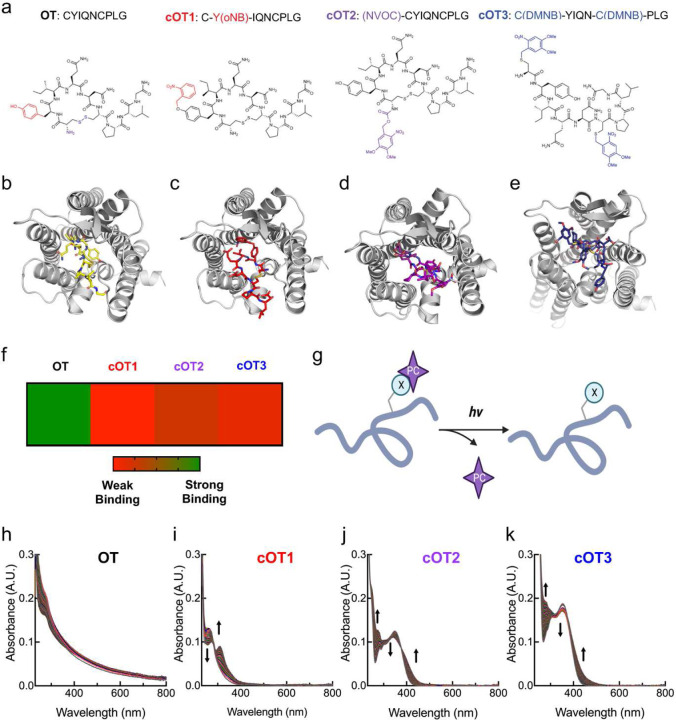
Figure 1. Structure, molecular modeling, and photophysical properties of oxytocin and caged oxytocin derivatives.
(a) Amino acid sequences and chemical structures of oxytocin (OT) and the photocage-modified oxytocin analogs cOT1, cOT2, and cOT3. For the OT structure the functional groups that are modified with cages are color-coded: the Tyr residue is labeled in red, the N-terminus in purple, and the disulfide in blue.
(b) Structure of OT bound to the oxytocin receptor, adapted from PDB: 7RYC.
(c-e) Structure of photocaged-modified oxytocin analogs cOT1 (c), cOT2 (d), and cOT3 (e) bound to the oxytocin receptor (PDB: 7RYC) through molecular docking modeling.
(f) Heat map analysis of OT, cOT1, cOT2, and cOT3 screened against the oxytocin receptor (PDB: 7RYC) via molecular docking modeling. In the gradient ruler, green color indicated strong binding, while red color indicates weak binding. Results listed from weakest binder: cOT1, cOT3, and cOT2.
(g)General reaction scheme of uncaging with UV light to yield oxytocin from caged oxytocin; X: caged residue, PC: photocage.
(h-k) Changes in the absorption spectrum of OT (h), cOT1 (i), cOT2 (j), and cOT3 (k) upon irradiation with 365 nm light (20 μM, room temperature, 1% DMSO/PBS). The spectra of each irradiated caged analog (cOT1, cOT2, and cOT3) show a significant change in absorbance, corresponding to photo-uncaging and oxytocin release.