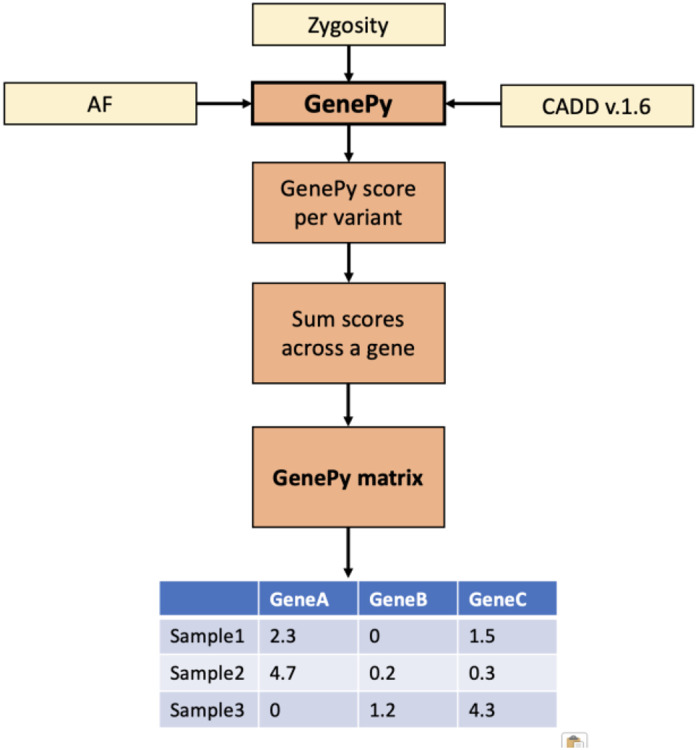
Figure 1 ∣. Overview of GenePy pathogenicity software and output.
GenePy takes input from an annotated variant call file and uses allele frequency (such as defined by gnomAD), allele zygosity, and a deleterious metric, such as CADD. The GenePy software then scores each variant according to the GenePy equation. GenePy scores are summed per gene (although the user can also specify per exon, or per gene pathway if required). The resultant GenePy score (summed across a gene/region) is then represented in a GenePy matrix, with samples along the Y axis and gene/region along the X axis.