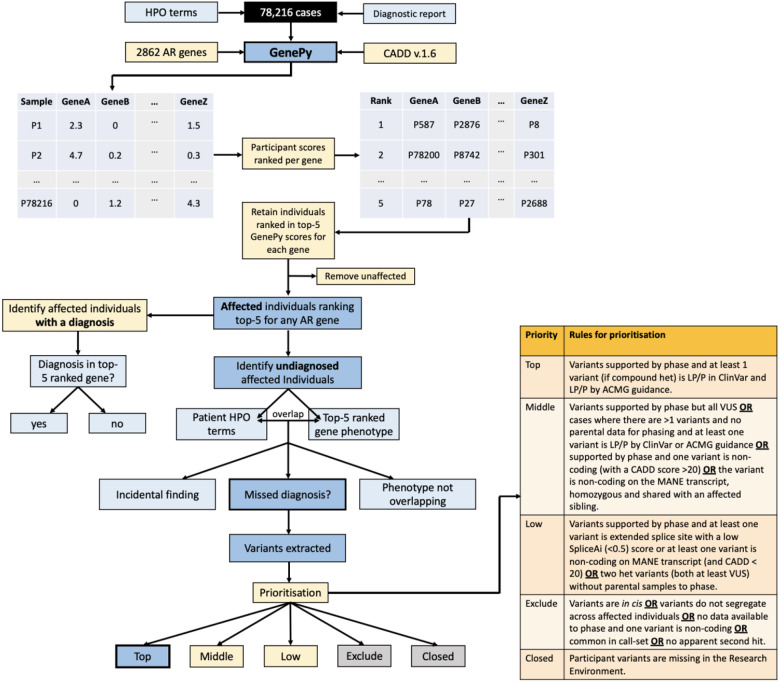
Figure 2 ∣. Workflow of GenePy applied to 78,216 cases in the 100,000 Genomes Project.
GenePy scores were created for 2862 autosomal recessive genes in 78,216 participants, using CADD v.1.6 and gnomAD v.2.1.1. Participants scores were ranked across the cohort per gene, whereby those who ranked in the top 5 GenePy score for each gene were retained for downstream analysis. Unaffected individuals were removed. HPO terms from unaffected individuals without a diagnosis returned by the 100,000 Genomes Project were compared with the clinical features described for the autosomal recessive gene that the participant scored in the top 5 for. If the participant’s HPO terms overlapped with the gene that the person ranked in the top 5 for, we extracted the individual participant variants and assessed phase, ClinVar status, and applied ACMG guidelines. We then prioritised the findings according to the prioritisation rules, with ‘Top’ priority. being putative missed diagnoses, ‘Middle’ and ‘Low’ priority being of interest but lacking sufficient evidence, ‘Exclude’ being not diagnostic and ‘Closed’ being when the participants had been withdrawn from the Project.