Abstract
Phase separation, in which macromolecules partition into a concentrated phase that is immiscible with a dilute phase, is involved with fundamental cellular processes across the tree of life. We review the principles of phase separation and highlight how it impacts diverse processes in the fungal kingdom. These include the regulation of autophagy, cell signaling pathways, transcriptional circuits and the establishment of asymmetry in fungal cells. We describe examples of stable, phase-separated assemblies including membrane-less organelles (MLOs) such as the nucleolus, as well as transient condensates that also arise through phase separation and enable cells to rapidly and reversibly respond to important environmental cues. We showcase how research into phase separation in model yeasts, such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, in conjunction with that in plant and human fungal pathogens, such as Ashbya gossypii and Candida albicans, is continuing to enrich our understanding of fundamental molecular processes.
INTRODUCTION
Organelles are often bound by a lipid membrane that separates internal components from the surrounding environment, but over the past decade multiple compartments have been identified in which macromolecules are concentrated in the absence of a phospholipid membrane. Such membrane-less organelles (MLOs) form by phase separation which involves de-mixing of a super-saturated solution into a dense phase that exists together with a more dilute surrounding phase1. Many MLOs have liquid-like behaviour consistent with liquid-liquid phase separation (LLPS), and their components can undergo rapid exchange with the surrounding environment2,3. The physics of polymer systems undergoing phase separation have been well studied through thermodynamic models such as the Flory-Huggins theory4. When a polymer is mixed with a solvent, phase separation can occur above a crucial concentration that depends on environmental factors (temperature, osmolarity, and pH) that alter the effective favourability (free energy) of polymer-polymer interactions. MLOs that form via phase separation take several forms, including liquid-like assemblies (via LLPS), semi-solid gels (with viscoelastic behavior) and rigid fibrillar aggregates, and are associated with diverse cellular processes throughout the fungal kingdom. A primer on the molecular forces underlying phase separation is provided in BOX 1.
Box 1. Molecular forces promoting phase separation.
Multivalent interactions, in which a single component contacts multiple other components, are critical drivers of phase separation. Multivalency can be achieved by proteins with intrinsically disordered regions (IDRs) or can involve interactions between folded domains and short linear motifs (SLiMs) that are sequence-specific recognition sites1,3,106–109 . Prion-like domains (PrLDs) are a particularly important class of IDRs and are defined by their sequence composition which is enriched in uncharged polar amino acids and glycine similar to S. cerevisiae prions.
A “sticker and spacer” model has been developed to describe how phase-separating IDRs interact. This simplified model defines “sticker” residues as those involved in electrostatic, hydrophobic, cation–Pi or Pi–Pi interactions, while “spacer” residues do not contribute to intermolecular interactions109,110. Attention has focused on aromatic residues as important “stickers” that can form Pi-Pi interactions with other aromatic residues or cation-Pi interactions with basic residues107,109–111. The nature of the spacer residues also defines biocondensate properties with glycine spacers promoting liquid behavior whereas glutamine/serine spacers promote solidifying of condensates110.
Overall, many residue types and intermolecular forces can contribute to phase separation, such as hydrophobic contacts/amino acids that can drive condensation of IDRs108,112,113. Formation of transient structures (including β-sheets and intermolecular helical regions) can also increase self-assembly and condensation formation114–116. In fact, one model proposes that transient structures are intermediate events in the formation of labile β-sheet structures that drive LLPS116–118, although this model is contested113,119.
Phase-separating molecules have further been designated as “scaffolds”, that are both necessary and sufficient for phase separation, and “clients”, which can selectively partition into condensates but do not phase separate by themselves120,121. Condensation also can be coupled to the formation of a system-spanning or “percolation” interaction network, although percolation networks can also occur independently of phase separation122.
Box 1 Fig. Weak protein-protein interactions and multivalency drive phase separation.
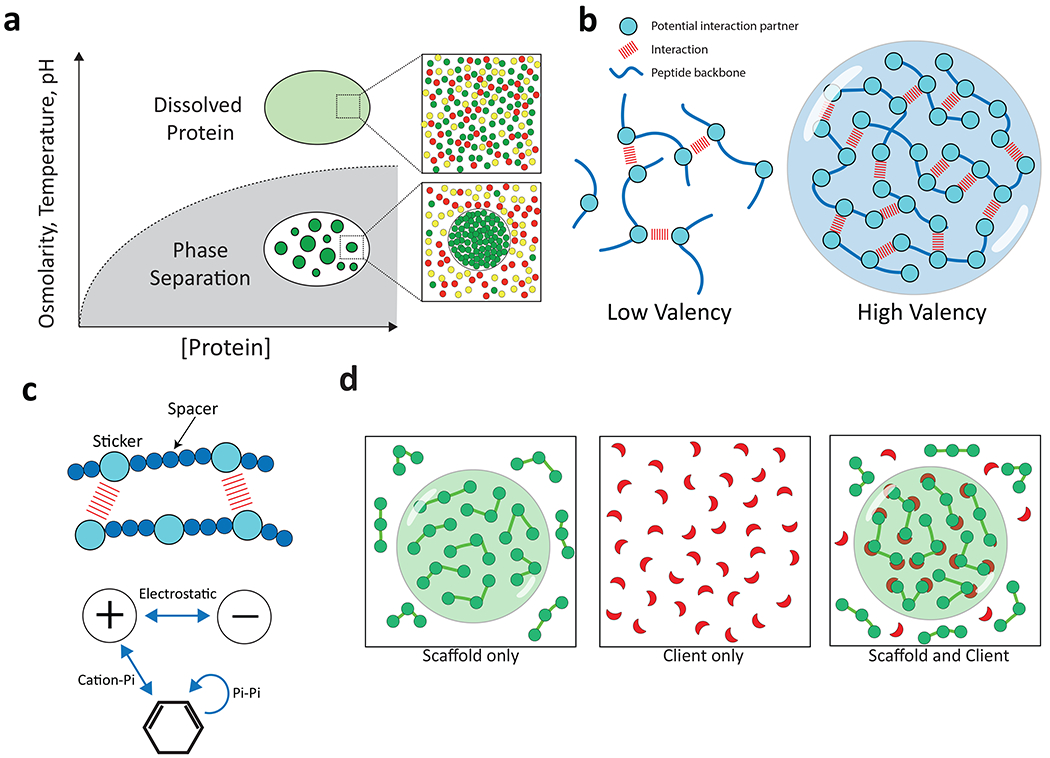
a, Phase separation occurs when protein concentration, osmolarity, temperature, and/or pH cross a threshold where intermolecular interactions drive assembly into a dense phase that co-exists with the surrounding dilute phase (adapted from ref 123). b, Higher valency due to a higher number of potential interactions between two peptide chains promotes phase separation. c, A “sticker” and “spacer” model for phase separation. Charged and aromatic “sticker” residues (larger balls) are distributed along a polypeptide interspersed with stretches of polar, hydrophilic residues (smaller balls) that act as “spacer” residues. d, “Scaffold” proteins have the capacity to undergo phase separation independent of other factors due to their high valency. Scaffolds can recruit “client” proteins that by themselves are not able to undergo phase separation under the same conditions.
Here, we review the diverse roles of biomolecular condensates in model and pathogenic fungi, including their function in autophagy, cytoskeletal organization and cell polarity, transcriptional regulation of cell fate, and sensing and responding to the cellular environment.
Stress-induced bodies formed by phase separation
Stress granules (SGs) and processing (P) bodies are dynamic MLOs that assemble on mRNAs stalled in translation (FIG. 1). Different SGs can form depending on the stress, which leads to the release of mRNAs encoding heat shock and chaperone factors thereby enabling their translation and stress adaptation5. P bodies are constitutively present in the cytoplasm and share components with SGs, but do not include translation initiation factors and instead sequester proteins associated with mRNA decay6. SGs and P bodies therefore perform distinct functions, and these MLOs can also be found docked against one another with mRNAs moving between them7. In S. cerevisiae, increased temperature or depletion of specific nutrients triggers SG formation and is driven by high concentrations of IDRs forming on mRNPs following stalled translation8. This causes the assembly of condensates that then mature into a less dynamic, stable core that is surrounded by a liquid shell8. Both yeast and mammalian SGs share a similar architecture, although a more extensive solid core in yeast SGs makes them less dynamic overall8,9.
Figure 1. Stress granule and P body formation in response to environmental changes.
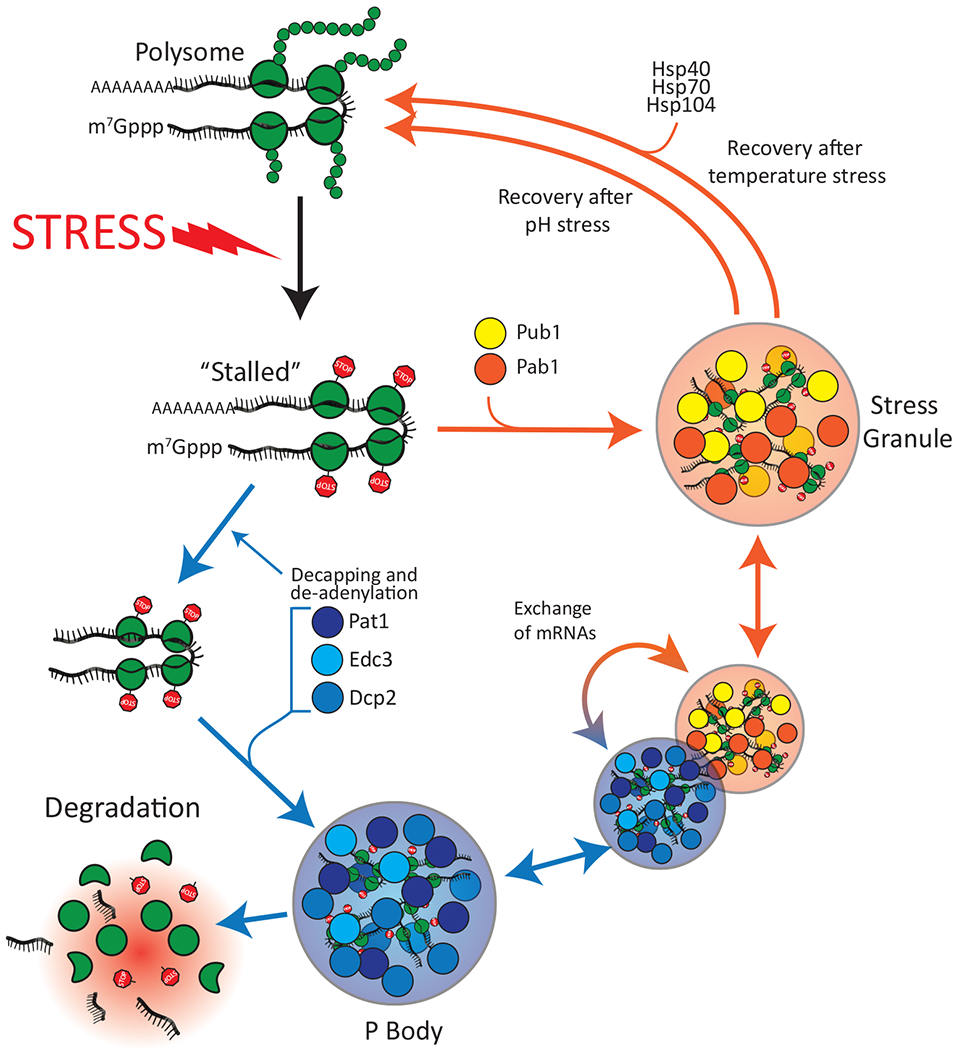
Extracellular stress such as heat shock or a sudden drop in pH leads to global inhibition of translation and ribosome stalling. Stalled mRNAs are diverted to either P bodies or stress granules. Following pH stress, stress granules dissolve spontaneously, whereas stress granules formed following temperature stress require the assistance of Hsp40, Hsp70 and Hsp104. Subsets of mRNAs that are concentrated in P bodies can either be degraded or exchanged with stress granules.
S. cerevisiae poly(A)-binding protein Pab1 is a canonical SG factor that contains four RNA recognition motifs (RRMs) as well as a proline-rich disordered domain (P domain)5,10. Pab1 phase separates in vitro to form gels in response to physiological cues such as a reduction in pH or heat shock5. RRMs promote Pab1 phase separation through electrostatic interactions, while hydrophobic interactions between P domains enhance this process5. A second S. cerevisiae SG protein, Pub1 (poly(U)-binding protein), is similar to Pab1 in that RRMs drive self-assembly while IDRs modify condensate properties11. Notably, Pub1 condensates show distinct material states depending on the stress; purified Pub1 forms reversible, gel-like condensates in response to low pH whereas more solid-like structures arise following heat shock, replicating observations with Pub1-containing condensates in cells11. Interestingly, only the more solid, thermally-induced condensates require the Hsp104 chaperone for dissolution11 (FIG. 1), indicating differences in the structural architecture of these condensates. Heat-induced condensates formed by Pab1 are similarly dispersed by an active disaggregation system consisting of Hsp40/Hsp70/Hsp104 chaperones which therefore helps cells recover from heat shock12.
P bodies function in RNA metabolism including mRNA storage/decay, and are also induced by stress13. These bodies are archetypal condensates with liquid-like properties both in S. cerevisiae and in mammals, and several purified P body factors have been shown to undergo LLPS in vitro9,14,15. Multivalency in P bodies is achieved by interactions between folded protein domains, IDRs, and RNA14. The most highly enriched P body proteins in S. cerevisiae (Dcp2, Pat1, and Edc3) partition cooperatively into these bodies as well as promote P body assembly16 (FIG. 1). Importantly, these studies suggest that only a few factors need to evolve LLPS capacity in order for MLOs to form16. In heavily stressed cells, the liquidity of P bodies is maintained by Hsp104 and loss of this activity results in P body proteins entering into SGs, further highlighting how disaggregases determine the behavior of stress-induced MLOs in the cell9.
A third class of stress-induced condensate in S. cerevisiae is the glycolytic (G) body, which involves glycolytic enzymes assembling into gel-like condensates during hypoxia17,18. G bodies enable growth under hypoxic conditions when oxidative phosphorylation is unavailable, most likely by concentrating glycolytic enzymes within condensates and promoting glucose consumption18. G body formation involves multivalent protein-protein and protein-RNA interactions and, as with other MLOs, RNA may act as a scaffold for development of these bodies17. Additional factors recruited to G bodies include Hsp70 chaperones (Ssa1/Ssa2) and the AMP-activated protein kinase Snf1p, with the latter necessary for G body formation18. Analogous G-like bodies are present in human hepatocarcinoma cells under hypoxic stress and where RNA enables the formation of these metabolic bodies18, suggesting conservation across eukaryotes.
Changes to the cellular environment that drive phase separation
Upon depletion of energy, S. cerevisiae and S. pombe yeast cells enter into a state of dormancy associated with cytoplasmic acidification19,20. The drop in intracellular pH occurs because culture medium is acidic whereas the intracellular pH is neutral, and cells must expend energy to maintain this pH difference21,22. Energy depletion and intracellular acidification cause a number of cytoplasmic proteins to assemble into microscopically visible foci or filaments19,23. Under these conditions, the yeast cytoplasm also transitions from its normal fluid state to a more solid-like state, which may be because much of the yeast proteome becomes less soluble and forms higher order assemblies19,20,24. This response is not unique to yeast cells as bacterial cells similarly respond to glucose starvation by transitioning from a glassy-liquid to a solid-like state, suggesting that these transitions are conserved and promote adaptation to stress20,25.
The S. cerevisiae translation termination factor Sup35, long studied for its ability to form a heritable prion, also forms condensates upon a drop in intracellular pH26. Sup35 consists of an N-terminal prion-like domain (PrLD; see BOX 1), a negatively charged middle (M) domain, and a C-terminal GTPase domain that catalyzes translation termination. Franzmann et al. showed that the M-domain acts as a pH sensor and causes Sup35 to form gel-like droplets following a stress-induced pH reduction, and that droplets redissolve upon restoration of pH, both in vitro and in cells26. In contrast, the Sup35 C-terminal domain forms irreversible aggregates during stress when expressed alone, as the PrLD is necessary for preventing aggregate formation. Reversible gel formation appears to be the ancestral role of Sup35, as the S. pombe ortholog cannot propagate as a prion but shares the ability to form stress-induced condensates26. Thus, most PrLDs (S. cerevisiae contains >200 proteins with such domains) likely function to modulate phase separation or protein solubility rather than act as prions, with the latter being the exception rather than the rule27. Debate continues as to the relative importance of S. cerevisiae Sup35 in forming condensates versus the prion state, with both roles being potentially beneficial to this species28.
Related studies have demonstrated that increased macromolecular crowding can promote phase separation events. Crowding agents such as polyethylene glycol have been extensively used to increase effective protein concentrations and promote phase separation in vitro29. To examine how macromolecular crowding influences behavior within cells, Delarue et al. performed microscopic tracking of genetically encoded multimeric (GEM) nanoparticles30. GEMs consist of a fluorescent molecule fused to a scaffold which multimerizes into particles of a defined shape and size. When expressed in S. cerevisiae cells, the motility of GEMs was reduced in the relatively crowded nucleus compared to that in the cytoplasm31 (FIG. 2a). Using this system, increased mTORC1 activity was shown to increase macromolecular crowding due to an increase in ribosome number30 (FIG. 2b). Ribosomes are a major cellular component (~200,000 ribosomes are present per yeast cell) and occupy ~20% of the cytosolic volume; an increase in the number of ribosomes therefore increased both macromolecular crowding and phase separation of a cytosolic protein30. These experiments establish close links between ribosome concentrations, macromolecular crowding and phase separation in yeast, with similar results obtained in a human cell line30. A recent study showed that increased macromolecular crowding also occurs in energy-starved S. cerevisiae cells due to a reduction in cell size, which in turn supports the formation of MLOs24.
Figure 2. The effect of molecular crowding on phase separation.
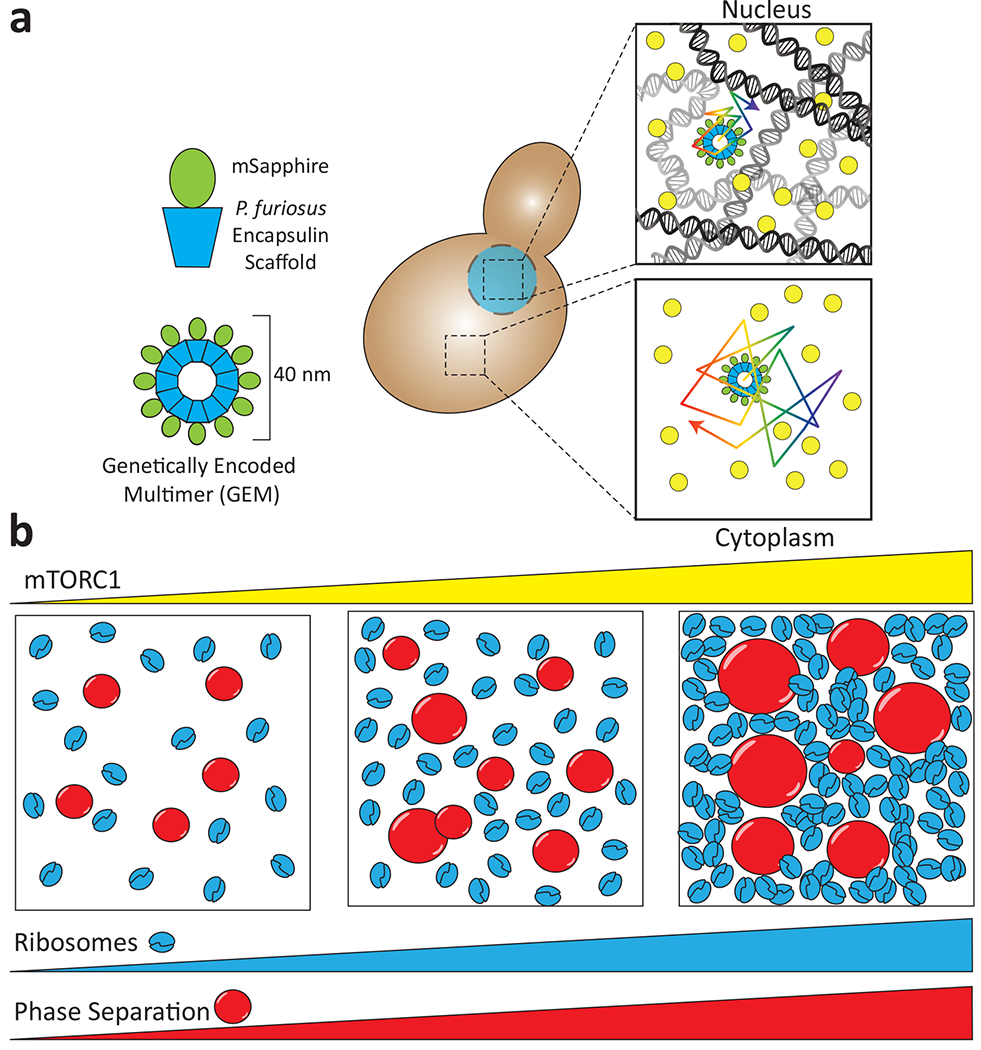
a, Microrheology using self-assembling Genetically Encoded Multimers (GEMs) allows measurement of intracellular crowding in the nucleus and the cytoplasm. Individual monomers consist of a Pyrococcus furiosus encapsulin scaffold fused to a fluorescent protein and spontaneously assemble into 40 nm spheres. The nucleus is a more crowded milieu than the cytoplasm and thus the random thermal motion of GEMs is decreased. b, mTORC activation following starvation results in an increased ribosome number. This increase in molecular crowding can increase phase separation of cytoplasmic proteins. Adapted from 30.
Taken together, these studies reveal that starvation can drive phase separation and oligomerization of cellular factors due to both changes in intracellular pH and increased molecular crowding. Recent experiments further show that hyperosmotic stress can also drive the formation of intracellular protein foci (OSF; osmotic shock foci) which may represent liquid droplets formed upon increased intracellular crowding32. Given that biocondensates arise in response to multiple stresses, it is important to note that protein constituents can be shared between different stress-induced condensates. For example, Sup35 and the SG protein Pab1 partially colocalize in pH-stressed cells but not in starvation-stressed cells26. A key ongoing research question is to therefore determine what controls the targeting of molecules to different stress-induced condensates in the cell.
The cytoskeleton, cell polarity and control of nuclear division
The cytoskeleton is a dynamic structure composed of filaments that undergo nucleation, polymerization, and depolymerization. In both budding yeast and filamentous fungi, the polarisome nucleates actin polymerization which involves the formin protein Bni1 together with nucleation promoting factor (NPF) and the scaffold protein Spa233,34. In S. cerevisiae, polarisome proteins concentrate at the bud tip via LLPS to nucleate actin assembly while remaining in exchange with the surrounding cytoplasm34,35. Xie et al. identified actin-interacting protein 5 (Aip5) as a factor that synergistically promotes actin assembly with Bni134. Intriguingly, the N-terminal domain of Aip5 is an IDR that causes the formation of amorphous condensates in vitro, while addition of Spa2 to Aip5 assemblies turns these condensates into more dynamic, liquid-like droplets34,35. It is therefore envisaged that Spa2 prevents Aip5 aggregation during stress, which in turn enables condensate dissolution and supports the restart of actin assembly during recovery from stress34,35 (FIG. 3a).
Figure 3. Phase separation in polarized growth, cell asymmetry and nuclear divisions.
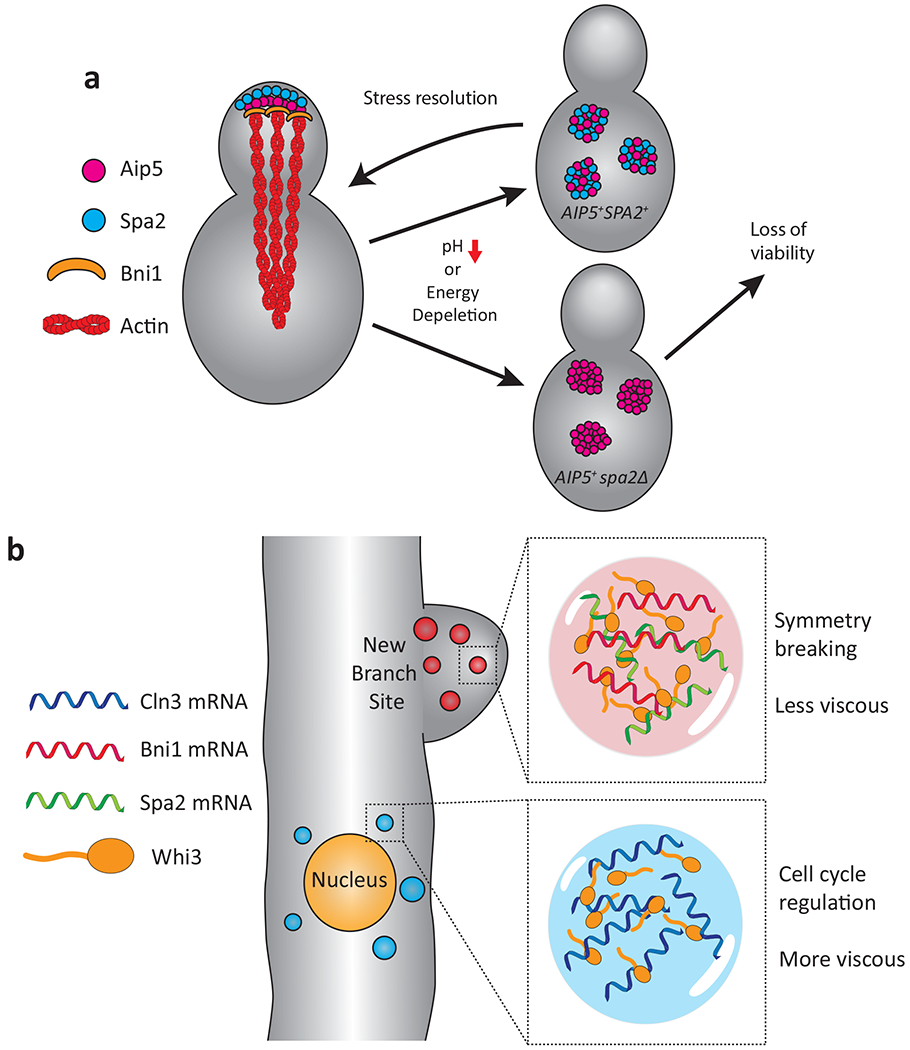
a, Spa2 localizes Aip5 to the S. cerevisiae bud tip, recruiting Bni1 and nucleating actin filaments. Stresses such as low pH or energy depletion result in Aip5 and Spa5 forming cytoplasmic condensates. When both Aip5 and Spa2 are present, these condensates are rapidly disassembled following removal of the stress. In the absence of Spa2, Aip5 forms more stable condensates that are not readily disassembled and there is a consequent loss of viability following prolonged stress. Adapted from 34. b, In A. gossypii, mRNA binding protein Whi3 forms distinct protein/mRNA condensates. Whi3 condensates formed with Bni1and Spa2 mRNA are localized to the site of branch formation (symmetry breaking) where they nucleate actin assembly. Whi3 droplets containing Cln3 mRNA form adjacent to the nucleus where they regulate asynchronous nuclear division (cell cycle regulation). Adapted from 36.
Ashbya gossypii is a filamentous fungus that is a plant pathogen and, like S. cerevisiae, belongs to the family Saccharomycetaceae. A. gossypii has emerged as an important model organism for studying cell polarity, filamentation and how asynchronous nuclear divisions occur within multinucleated cells. Remarkably, the protein Whi3 has been linked to the regulation of both cell polarity and the timing of nuclear divisions due to its ability to form RNA-dependent liquid droplets36–38 (FIG. 3b). Whi3 establishes polarity at symmetry breaking points in the cell by forming condensates with Puf2 that incorporate BNI1 and SPA2 RNA transcripts36,37. In contrast, Whi3 droplets near nuclei contain CLN3 mRNA (encoding for a G1 regulatory cyclin), and differences in the spatial distribution of this transcript determine the timing of the nuclear divisions in multinucleate cells36,38. Zhang et al. showed that the presence of different mRNAs results in distinct types of Whi3 condensates, from more liquid-like to more gel-like assemblies36. These results therefore provide a striking example of how RNA can impact the physical properties of biocondensates and can result in changes in their size, shape, viscosity, surface tension and composition36,39 (also see BOX 2 for the central role of nucleic acids in promoting phase separation).
Box 2. Protein-nucleic acid interactions drive condensate formation.
Negatively charged nucleic acids can interact with positively charged proteins to drive phase separation, a form of complex coacervation where multivalent nucleic acid-binding domains can support condensate formation even in the absence of IDRs124. Single-stranded nucleic acids can also participate in cation-pi or pi-pi interactions through exposed aromatic bases thereby supporting condensate formation, while recognition of nucleotide bases via hydrogen bonds is an additional driving force for phase separation125.
RNA is a critical component of multiple condensates including stress granules, storage granules, and various speckles, paraspeckles, nuclear speckles and transcriptional complexes. RNA transcripts can directly seed the nucleation of condensates or can induce protein conformations that promote condensation126,127. RNA levels can also tune phase separation of ribonucleoprotein (RNP) condensates. For example, low RNA levels stimulate condensate formation by human FUS protein while high RNA levels suppress condensate formation, and changes in RNA concentrations may underlie aberrant LLPS structures and pathologic assemblies128. While liquid condensates adopt a spherical shape in the absence of external forces, certain RNP-containing condensates form highly viscous structures, mesh-like assemblies or filamentous networks depending on the RNA substrates involved, highlighting how various shapes and structures can arise via phase separation39,129.
Autophagy
Autophagy is the organized breakdown of components by the lysosome, and provides energy and building blocks for cells to survive stress. This process involves formation of the Pre-Autophagosomal Structure (PAS) on the cytoplasmic face of the vacuolar membrane, which nucleates assembly of a cup-shaped isolation membrane adjacent to the PAS that engulfs material in an autophagosome40,41. S. cerevisiae PAS formation involves five IDR-containing Atg (autophagy-related) proteins that co-assemble into the ATG1 complex (FIG. 4a)40–42. In starved cells, the TORC1 kinase is inactivated and Atg13 becomes dephosphorylated (by PP2C phosphatases) and establishes multivalent interactions with Atg17 to form highly liquid droplets42,43. Atg13 also interacts with the Vac8 membrane protein to anchor ATG1 droplets at the vacuolar membrane where they fuse to form one large condensate – the PAS40,42. Within the PAS, Atg1 is activated by autophosphorylation and phosphorylates Atg13, creating an equilibrium between phospho-Atg13 in the dilute phase and its unphosphorylated form in the condensed phase, which is important for maintenance of the PAS42 (FIG. 4a). Formation of the cupped isolation membrane is not fully understood but is initiated by the recruitment of Atg9-containing vacuoles to the PAS via interactions with Atg13, and these vacuoles then fuse to generate the isolation membrane44,45 (FIG. 4a). ATG proteins are conserved and phase separation is likely to play related roles in autophagy in multicellular organisms and yeast, although autophagosome formation is organized differently (both spatially and temporally) between these species41.
Figure 4. Phase separation regulation of bulk and selective autophagy.
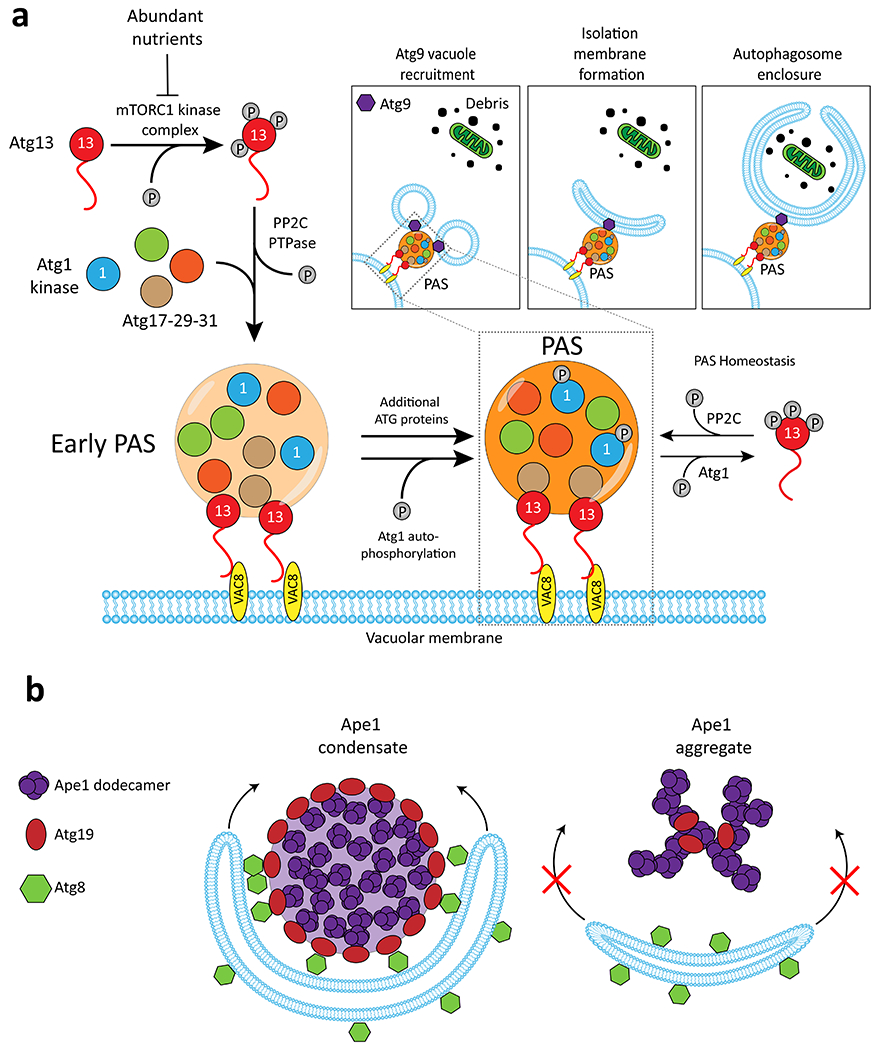
a, When nutrients are abundant, Atg13 is hyperphosphorylated by TORC1 kinase. Upon nutrient starvation, Atg13 is dephosphorylated by PP2C phosphatase enabling interactions with the Atg17-29-31 complex and the Atg1 kinase which then coalesce into condensates. The resulting Pre-Autophagosomal Structure (PAS) is tethered to the vacuolar membrane by interactions with Vac8. In the newly formed PAS, Atg1 auto-phosphorylates itself and hyper-phosphorylates Atg13. Cytoplasmic phospho-Atg13 is trafficked back to the PAS following dephosphorylation by PP2C. Thus, Atg1 and PP2C maintain an equilibrium of unphosphorylated and phosphorylated Atg13 which maintains the PAS. The PAS is also the site of Isolation Membrane formation, which in turn will become the mature autophagosome. Inset, Following PAS formation, vacuoles containing ATG9 are recruited and subsequently fuse to become the cupped isolation membrane, which grows to engulf materials destined for destruction in the lysosome. b, Dodecamers of Ape1 undergo phase separation and are degraded by selective autophagy in nutrient rich conditions. The Ape1 condensate is coated by a shell of the Atg9 adapter protein which templates growth of Atg8-decorated isolation membrane. Ape1 mutants which form aggregates are not engulfed. Adapted from 47.
A distinct form of autophagy, selective autophagy, targets specific organelles and biomolecules to the vacuole even under nutrient-rich conditions and also involves phase separation. S. cerevisiae Ape1 is the principle cargo of a form of selective autophagy termed the cytoplasm-to-vacuole targeting (Cvt) pathway41,46. Ape1 contains an N-terminal propeptide that can form a helical structure and self-assemble into dodecamers that then coalesce into semi-liquid droplets46–48. Atg19 acts as a receptor for Ape1 as it “floats” on the surface of Ape1 condensates and connects these condensates to Atg8/Atg21 (Fig. 4b)46–48. Through these interactions, a shape change in the isolation membrane enables it to form a phospholipid bilayer around Ape1 droplets and sequester them for degradation47. Changing a single amino acid in the Ape1 propeptide produced amorphous aggregates rather than gel-like droplets, and these hardened structures failed to interact with Atg19/Atg8 or undergo autophagy47. Similar results have been observed in the targeting of Caenorhabditis elegans P granule proteins for autophagic degradation, where the gel-like state provides a suitable platform for engulfment by autophagosomal membranes49. These results highlight how the material properties of condensates are critical for determining the destination of cellular cargoes, both in fungi and higher eukaryotes.
Regulation of autophagy via sensing of reactive oxygen species
S. cerevisiae cells switched from a nutrient-rich medium to a minimal medium containing a non-fermentable carbon source undergo autophagy even in the continued presence of nitrogen, which can help cells maintain mitochondrial health during respiratory growth50. Tu and colleagues revealed that reactive oxygen species (ROS) produced by high mitochondrial dysfunction under these conditions are sensed by Pbp1, the yeast ortholog of mammalian ataxin-251,52. Remarkably, Pbp1 is capable of undergoing phase separation in a redox-sensitive manner; this protein readily forms liquid- or gel-like droplets in vitro but these droplets dissolve upon the addition of hydrogen peroxide51. This mechanism involves oxidation of methionine residues within the Pbp1 LCR which therefore acts as a reversible readout of mitochondrial respiratory status51. Thus, Pbp1 condensates form during high respiratory growth and are poised to sense mitochondrial dysfunction and increased ROS levels, which in turn results in activation of TORC1 and inhibition of autophagy (as a means of adaptation to mitochondrial stress)52,53. These studies provide a striking example of how phase separation can be tuned by key physiological signals such as ROS. It was subsequently shown that human TDP-43 (involved in the formation of neuronal granules) similarly forms redox-sensitive condensates54, highlighting how observations in S. cerevisiae led to the discovery of ROS sensing via phase separation in higher organisms.
Phase separation as a mechanism for sensing carbon dioxide
Cells have evolved sensitive mechanisms to detect changes in the levels of carbon dioxide (CO2). In fungi, CO2 can affect multiple processes including mating, meiosis, phenotypic switching and filamentation55. Zhang et al. revealed that CO2 sensing in the opportunistic human fungal pathogen Candida albicans involves condensates formed by Ptc2, a member of the PP2C family of phosphatases56. Here, a serine/threonine-rich region within the IDR of Ptc2 enabled this protein to undergo CO2-induced phase separation, with CO2 envisaged as a “molecular glue” that bridges interactions between Ptc2 molecules thereby stimulating condensate formation56. A related PP2C phosphatase from plants showed a similar ability to undergo CO2-induced phase separation56, indicating that this mechanism of environmental sensing (while uncovered in fungi) is likely conserved across diverse eukaryotic species56.
Nuclear compartmentalization and heterochromatin
Phase separation plays a central role in the establishment of nuclear compartments such as the nucleolus which houses ribosome biogenesis and ribonucleoprotein assembly. This organelle is organized into three nested sub-compartments in higher eukaryotes; a core fibrillar center (FC) exists inside a dense fibrillar component (DFC), which itself resides within the granular component (GC)57. These compartments represent coexisting, immiscible liquid phases – this layered, multiphase architecture was reproduced in vitro as coincubation of a DFC protein with a GC protein generated multiphase droplets57. Here, IDRs within these proteins drive phase separation while RNA binding domains contribute to the immiscibility of the protein phases57. S. cerevisiae nucleoli have only two sub-compartments58 and modeling suggests that ribosomal DNA (rDNA) is phase separated from bulk chromatin due to crosslinks between rDNA repeats59, while nucleolar RNPs also undergo phase separation60. Indeed, the combination of rDNA phase separation and tethering of rDNA repeats to the nuclear envelope may explain the characteristic crescent shape of the yeast nucleolus61.
Phase separation also controls the formation of transcriptionally repressed heterochromatin. Pioneering work on heterochromatin protein 1 (HP1) in higher eukaryotes showed that it can undergo phase separation and recruit other heterochromatin-associated factors involved in transcriptional repression62,63. Constitutive heterochromatin is marked by trimethylation of lysine 9 on histone H3 (H3K9me3), and interactions between these marks and both HP1 chromodomains and SUV39H1 (which introduces H3K9me3 marks) are drivers of phase separation64. DNA polymers also contribute to the properties of heterochromatin resulting in stable structures that resist mechanical forces, while HP1 can exchange between condensate and non-condensate populations65. Swi6, the S. pombe homolog of HP1, similarly undergoes phase separation with reconstituted chromatin that contains H3K9me3 marks, suggesting parallels between constitutive heterochromatin formation in yeast and humans64. Related mechanisms may establish the formation of facultative heterochromatin in which polycomb proteins form phase-separated condensates and are associated with chromatin that is trimethylated on lysine 27 of histone H3 (H3K27me3)66,67.
Phase separation, transcriptional activation and super-enhancer-like elements
Multiple studies have observed that transcription factors (TFs) can assemble into phase-separated complexes to activate gene expression68–72. Phase separation may occur preferentially at mammalian super-enhancers (SEs), where high concentrations of TFs, Mediator complex and RNA polymerase II (Pol II) span genomic regions >10 kb69,70,72 (FIG. 5a,b). Weak multivalent interactions between TFs and coactivators together with structured interactions between TFs and DNA promote phase separation70. The sharply defined thresholds associated with condensate formation may underlie unique SE properties such as their hypersensitivity to changes in TF levels69. Moreover, the de novo assembly of a SE can be initiated by binding of a single TF to an enhancer, as exemplified by somatic mutations that cause MYB binding and subsequent TAL1 overexpression in T-cell acute lymphoblastic leukemia73.
Figure 5. Phase separation regulates transcription and fungal cell fate.

a, Coordinated binding of multiple TFs to regions upstream of their ORFs is often observed even without consensus binding sites for these regulators, suggestive of recruitment by protein-protein interactions. b, A phase separation model of transcription where TFs form condensates together with the transcriptional machinery to regulate the expression of cell identity genes. c, C. albicans switches epigenetically between “white” and “opaque” phenotypic states. d, The white-to-opaque transition is regulated by a TF network whose members bind to their own promoters as well as those of others in the network, as indicated by the arrows. Adapted from ref 82. e, RNA polymerase II interacts with transcriptional initiation or elongation condensates depending on the phosphorylation state of its C-terminal domain (CTD).
Mammalian SEs have been linked to transcriptional regulatory networks (TRNs) that define cell fate, where a core set of master TFs act in concert to control the expression of large numbers of target genes74,75. Intriguingly, fungal TRNs are also regulated by sets of master TFs operating at super-sized regulatory regions (“SE-like elements”). Prominent examples of fungal TRNs include those regulating pseudohyphal formation in S. cerevisiae, the temperature response in Histoplasma capsulatum, biofilm formation and phenotypic switching in C. albicans, and the heat shock response that is conserved from fungi to humans76–79. The TRN regulating white-opaque phenotypic switching in C. albicans has been extensively analyzed and involves eight master TFs that act together at SE-like elements80,81 (FIG. 5c,d). The master white-opaque TFs are often recruited to SE-like regions even in the absence of consensus DNA binding motifs, indicating they are likely recruited via protein-protein interactions. These regulatory TFs contain PrLDs that enable them to undergo phase separation in vitro, and mutations that block this process abolish their function, thereby linking phase separation to the regulation of this TRN82. Analysis of the heat shock response in S. cerevisiae similarly shows the presence of SE-like elements at which transcriptional condensates are expected to form83,84. Phase separation and SE-like regions are therefore implicated in transcriptional regulation in both fungi and mammals, as compared in TABLE 1.
Table 1. Comparison of super-enhancer features in mammalian cells with super-enhancer-like elements in C. albicans cells.
White-opaque and biofilm transcriptional regulatory networks (TRNs) are used as examples of SE-like regulatory regions in C. albicans.
| Super-enhancer features | Mammalian super-enhancers | C. albicans super-enhancer-like regions |
|---|---|---|
|
| ||
| Role in cell identity | Found to control cell identity and differentiation in murine embryonic stem cells (ESCs), multiple immune cell type, and to contribute to a broad range of cancers via enrichment at genes with oncogenic function. | Extended regulatory regions are required for white-opaque cell fate determination and additionally for control of biofilm formation, whereby cells transition between planktonic growth and communal growth. |
|
|
||
| Size | Median size is > 8 kb, whereas typical enhancers are ~700 bp. | For the white-opaque TRN, median size of upstream intergenic regions is > 7 kb, while average intergenic regions are ~ 557 bp. |
|
|
||
| TF enrichment levels | Elevated TF binding at constituent enhancers, increased cooperative transcriptional activation, and combined TF/coactivator enrichment ~10-fold higher than seen at typical enhancers. | Master TFs bind together at multiple positions across super-enhancer-like regions (see Fig. 5), although quantitative analysis of cofactor levels has not been performed. |
|
|
||
| Epigenetic marks | Relatively high levels of acetylation of histone H3 at lysine 27 (H3K27ac) are commonly used to define super-enhancers, sometimes in combination with other criteria. |
Unknown. |
|
|
||
| Sensitivity to TF perturbation | Highly sensitive – blocking binding of just one coactivator, like BRD4, can collapse entire super-enhancer. | Highly sensitive – a small increase or decrease in levels of the Wor1 TF, for example, can drastically alter white-opaque cell fate switching rates. |
|
|
||
Despite considerable interest in SEs it is currently unclear whether these elements are functionally distinct from regular enhancers, and the role of phase separation in transcription is also strongly debated85,86. Indeed, a recent study suggests that multivalent interactions can enable the formation of mammalian TF “hubs” that promote gene expression, yet when hubs assemble into larger, phase-separated condensates then gene expression is inhibited87. These results support a “Goldilocks” model whereby small changes in multivalent interactions can finely tune gene activation up or down, but where formation of condensates results in transcription inhibition87. Clearly, more work is required in this exciting area and examination of gene context will be critical, with condensates potentially enabling transcription at certain loci while restricting it at others. Additional technological innovations are therefore needed to further dissect the role of phase separation in transcription, including improved techniques to evaluate proteins by high resolution microscopy when expressed at endogenous levels in live cells.
Regulation of RNA polymerase II via phase separation
RNA Pol II contains a conserved, intrinsically disordered C-terminal domain (CTD) that enables it to form condensates, as well as to co-phase separate with transcription factors and transcriptional machinery88–91. The longer length of the mammalian CTD relative to the S. cerevisiae CTD (52 v. 26 heptad repeats) results in an increased propensity to undergo phase separation due to stronger CTD-CTD interactions88. CTD phosphorylation leads to dissociation of Pol II from Mediator condensates (associated with transcription initiation) and recruitment into condensates containing splicing factors91 (FIG. 5e). These transitions could involve direct maturation of Pol II-containing condensates or Pol II exiting initiation condensates before recruitment to splicing condensates92.
Related to this work, Quintero-Cadena et al. showed that Pol II CTD length correlates with gene density in eukaryotes, and proposed that this domain serves as a molecular bridge for Pol II to be recruited to active promoters, with longer CTD lengths enabling recruitment across greater distances93. CTD length also modulated transcriptional bursting with longer CTDs leading to stronger and more frequent bursts. Surprisingly, a truncated S. cerevisiae CTD was non-functional but function was restored by fusion to phase-separating IDRs from human FUS or TAF15 proteins93. Together, these studies demonstrate that CTD-CTD interactions, as well as CTD interactions with TFs, coactivators, and Mediator, assist the recruitment of Pol II for gene transcription.
RNA plays an integral role in transcriptional phase separation; low RNA levels increase the formation of Mediator condensates whereas high RNA levels dissolve these condensates94. This may result in a feedback mechanism whereby transcription dissipates condensates and causes the transcriptional bursts that are characteristic of this process94. Moreover, promoter-associated RNAs in mammalian cells are likely bound by multiple RBPs that enable Pol II and cofactors to reach the threshold levels necessary for phase separation95, and certain RBPs also promote Pol II release from initiation complexes to support transcription elongation96.
Recent studies in S. cerevisiae have similarly shown that TBP associated factor 14 (Taf14; a component of TFIID and chromatin remodeling complexes) utilizes multiple interaction partners to regulate transcription. Chen et al. demonstrated that a structured extra-terminal (ET) domain of Taf14 recruits co-factors via an ET-binding motif present on these partners97. Moreover, Taf14 formed condensates in vitro and binding partners could partition into these droplets, establishing that Taf14 can act as a scaffold to bring together co-factors and drive gene expression97. While the formation of phase-separated hubs is a recurring theme in transcription, Taf14 is, so far, an unusual example of a transcriptional regulator that is reported to use only structured domains to scaffold multi-component, phase-separated condensates97.
Fungi as model species to study amyloid disease
In addition to endogenous phase separation phenomena, fungi have been used to model the properties (and toxicities) of aggregative amyloids implicated in human disease98,99. In many cases, the precise role of condensate or amyloid formation in neurodegenerative diseases such as Huntington’s, amyotropic lateral sclerosis (ALS), Alzheimer’s and Parkinson’s is uncertain, although disease-causing mutations often increase amyloid formation in proteins associated with each disease100,101.
Several aggregation-prone human proteins have been analyzed in S. cerevisiae, with high-throughput screening used to identify new therapeutics that reduce amyloid toxicity102,103. Yeast have also been invaluable is in studying disaggregases that can detoxify aggregation-prone proteins associated with neurodegeneration. For example, S. cerevisiae Hsp104, or variants of this chaperone, can not only act as disaggregases (or anti-aggregation activities) on endogenous proteins but are also functional on human proteins associated with neurologic disorders despite metazoans lacking an ortholog of Hsp104104,105. Model fungi and yeast genetics therefore continue to be used to understand how aberrant phase transitions/aggregation can impact neurodegenerative diseases and to develop therapeutic interventions.
Conclusions
Phase separation and the formation of biomolecular condensates play central roles in virtually all aspects of biology, from ubiquitous cellular compartments to highly inducible assemblies. In fungi, the formation of condensates is highly sensitive to cellular conditions and phase separation therefore acts as an exquisite sensor of environmental cues, as evidenced in the responses to changes in pH, ROS and CO2 levels. Outstanding questions with regards to phase separation and MLOs in fungi include a better understanding of the molecular interactions that nucleate and stabilize phase separation; the contribution of transient or stable secondary structures to condensate formation; understanding the specificity by which proteins/nucleic acids are recruited to condensates; new tools to examine phase separation of proteins at endogenous levels in fungal cells; determination of which small molecules impact condensates; and developing therapeutic approaches including antifungal drugs based on understanding of phase separation biology. Studies in fungi will continue to be at the forefront of this field given the cell biological and genetic tools available in model fungi and the diversity of species being studied both as model organisms and as plant and human pathogens.
Acknowledgements
We would like to thank members of the Bennett lab for useful discussions and Dr. Benjamin Tu (UTSW) for feedback on sections of the review. Work in the Bennett lab is supported by NIAID grants AI141893/AI081704/AI166869 and work in the Fawzi lab is supported by NSF BIO 1845734 and NINDS R01NS116176.
Footnotes
Competing interests
NLF is a member of the scientific advisory board of Dewpoint Therapeutics.
References
- 1.Boeynaems S et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol 28, 420–435, doi: 10.1016/j.tcb.2018.02.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomes E & Shorter J The molecular language of membraneless organelles. J Biol Chem 294, 7115–7127, doi: 10.1074/jbc.TM118.001192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, doi: 10.1126/science.aaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Mitrea DM & Kriwacki RW Phase separation in biology; functional organization of a higher order. Cell communication and signaling : CCS 14, 1, doi: 10.1186/s12964-015-0125-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riback JA et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040 e1019, doi: 10.1016/j.cell.2017.02.027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov P, Kedersha N & Anderson P Stress granules and processing bodies in translational control. Cold Spring Harbor perspectives in biology 11, doi: 10.1101/cshperspect.a032813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon SL et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nature cell biology 21, 162–168, doi: 10.1038/s41556-018-0263-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain S et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498, doi: 10.1016/j.cell.2015.12.038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroschwald S et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807, doi: 10.7554/eLife.06807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson P & Kedersha N RNA granules. J Cell Biol 172, 803–808, doi: 10.1083/jcb.200512082 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroschwald S et al. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Reports 23, 3327–3339, doi: 10.1016/j.celrep.2018.05.041 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Yoo H, Bard JAM, Pilipenko EV & Drummond DA Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol Cell 82, 741–755 e711, doi: 10.1016/j.molcel.2022.01.005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y, Na Z & Slavoff SA P-Bodies: Composition, properties, and functions. Biochemistry 57, 2424–2431, doi: 10.1021/acs.biochem.7b01162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schutz S, Noldeke ER & Sprangers R A synergistic network of interactions promotes the formation of in vitro processing bodies and protects mRNA against decapping. Nucleic Acids Res, doi: 10.1093/nar/gkx353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromm SA et al. In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew Chem Int Ed Engl 53, 7354–7359, doi: 10.1002/anie.201402885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing W, Muhlrad D, Parker R & Rosen MK A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. eLife 9, doi: 10.7554/eLife.56525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller GG et al. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. eLife 9, doi: 10.7554/eLife.48480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin M et al. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell reports 20, 895–908, doi: 10.1016/j.celrep.2017.06.082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munder MC et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, doi: 10.7554/eLife.09347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyner RP et al. A glucose-starvation response regulates the diffusion of macromolecules. eLife 5, doi: 10.7554/eLife.09376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dechant R et al. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J 29, 2515–2526, doi: 10.1038/emboj.2010.138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orij R, Postmus J, Ter Beek A, Brul S & Smits GJ In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology (Reading) 155, 268–278, doi: 10.1099/mic.0.022038-0 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Petrovska I et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife, doi: 10.7554/eLife.02409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marini G, Nuske E, Leng W, Alberti S & Pigino G Reorganization of budding yeast cytoplasm upon energy depletion. Mol Biol Cell 31, 1232–1245, doi: 10.1091/mbc.E20-02-0125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry BR et al. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194, doi: 10.1016/j.cell.2013.11.028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzmann TM et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, doi: 10.1126/science.aao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Franzmann TM & Alberti S Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J Biol Chem 294, 7128–7136, doi: 10.1074/jbc.TM118.001190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyke DR, Dorweiler JE & Manogaran AL The three faces of Sup35. Yeast 36, 465–472, doi: 10.1002/yea.3392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andre AAM & Spruijt E Liquid-liquid phase separation in crowded environments. International Journal of Molecular Sciences 21, doi: 10.3390/ijms21165908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delarue M et al. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349 e320, doi: 10.1016/j.cell.2018.05.042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu T et al. nucGEMs probe the biophysical properties of the nucleoplasm. bioRxiv, 2021.2011.2018.469159, doi: 10.1101/2021.11.18.469159 (2022). [DOI] [Google Scholar]
- 32.Alexandrov AI et al. Analysis of novel hyperosmotic shock response suggests ‘beads in liquid’ cytosol structure. Biol Open 8, doi: 10.1242/bio.044529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140, 257–267, doi: 10.1016/j.cell.2009.12.031 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Xie Y et al. Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization. Nature Communications 10, 5078, doi: 10.1038/s41467-019-13125-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Y & Miao Y Polarisome assembly mediates actin remodeling during polarized yeast and fungal growth. J Cell Sci 134, doi: 10.1242/jcs.247916 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Zhang H et al. RNA controls polyQ protein phase transitions. Mol Cell 60, 220–230, doi: 10.1016/j.molcel.2015.09.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C, Occhipinti P & Gladfelter AS PolyQ-dependent RNA-protein assemblies control symmetry breaking. J Cell Biol 208, 533–544, doi: 10.1083/jcb.201407105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C et al. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell 25, 572–584, doi: 10.1016/j.devcel.2013.05.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden C & Gladfelter AS RNA contributions to the form and function of biomolecular condensates. Nature Reviews. Molecular Cell Biology 22, 183–195, doi: 10.1038/s41580-020-0264-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatogawa H, Suzuki K, Kamada Y & Ohsumi Y Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Reviews. Molecular Cell Biology 10, 458–467, doi: 10.1038/nrm2708 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Noda NN, Wang Z & Zhang H Liquid-liquid phase separation in autophagy. J Cell Biol 219, doi: 10.1083/jcb.202004062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujioka Y et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305, doi: 10.1038/s41586-020-1977-6 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto H et al. The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Dev Cell 38, 86–99, doi: 10.1016/j.devcel.2016.06.015 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto H et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198, 219–233, doi: 10.1083/jcb.201202061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki SW et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A 112, 3350–3355, doi: 10.1073/pnas.1421092112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki A & Noda NN Structural biology of the Cvt pathway. J Mol Biol 429, 531–542, doi: 10.1016/j.jmb.2017.01.003 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki A et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell 77, 1163–1175 e1169, doi: 10.1016/j.molcel.2019.12.026 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Yamasaki A et al. Structural basis for receptor-mediated selective autophagy of aminopeptidase I aggregates. Cell Reports 16, 19–27, doi: 10.1016/j.celrep.2016.05.066 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Zhang G, Wang Z, Du Z & Zhang H mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506 e1422, doi: 10.1016/j.cell.2018.08.006 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wu X & Tu BP Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell 22, 4124–4133, doi: 10.1091/mbc.E11-06-0525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M et al. Redox state controls phase separation of the teast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell 177, 711–721 e718, doi: 10.1016/j.cell.2019.02.044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang YS et al. Yeast ataxin-2 forms an intracellular condensate required for the inhibition of TORC1 signaling during respiratory growth. Cell 177, 697–710 e617, doi: 10.1016/j.cell.2019.02.043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prouteau M & Loewith R TOR signaling is going through a phase. Cell Metab 29, 1019–1021, doi: 10.1016/j.cmet.2019.04.010 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Lin Y et al. Redox-mediated regulation of an evolutionarily conserved cross-beta structure formed by the TDP43 low complexity domain. Proc Natl Acad Sci U S A 117, 28727–28734, doi: 10.1073/pnas.2012216117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin R, Pohlers S, Muhlschlegel FA & Kurzai O CO2 sensing in fungi: at the heart of metabolic signaling. Curr Genet 63, 965–972, doi: 10.1007/s00294-017-0700-0 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Zhang M et al. The intrinsically disordered region from PP2C phosphatases functions as a conserved CO2 sensor. Nature Cell Biology 24, 1029–1037, doi: 10.1038/s41556-022-00936-6 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Feric M et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697, doi: 10.1016/j.cell.2016.04.047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiry M & Lafontaine DL Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol 15, 194–199, doi: 10.1016/j.tcb.2005.02.007 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Hult C et al. Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res 45, 11159–11173, doi: 10.1093/nar/gkx741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrimore J et al. The rDNA is biomolecular condensate formed by polymer-polymer phase separation and is sequestered in the nucleolus by transcription and R-loops. Nucleic Acids Res 49, 4586–4598, doi: 10.1093/nar/gkab229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall AC, Ostrowski LA & Mekhail K Phase separation as a melting pot for DNA repeats. Trends Genet 35, 589–600, doi: 10.1016/j.tig.2019.05.001 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Larson AG et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240, doi: 10.1038/nature22822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245, doi: 10.1038/nature22989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell 76, 646–659 e646, doi: 10.1016/j.molcel.2019.08.019 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Keenen MM et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 10, doi: 10.7554/eLife.64563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eeftens JM, Kapoor M, Michieletto D & Brangwynne CP Polycomb condensates can promote epigenetic marks but are not required for sustained chromatin compaction. Nature Communications 12, 5888, doi: 10.1038/s41467-021-26147-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatavosian R et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem 294, 1451–1463, doi: 10.1074/jbc.RA118.006620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boija A et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 e1816, doi: 10.1016/j.cell.2018.10.042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A phase separation model for transcriptional control. Cell 169, 13–23, doi: 10.1016/j.cell.2017.02.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shrinivas K et al. Enhancer features that drive formation of transcriptional condensates. Molecular Cell 75, 549–561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415, doi: 10.1126/science.aar4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958, doi: 10.1126/science.aar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mansour MR et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377, doi: 10.1126/science.1259037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiter F, Wienerroither S & Stark A Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev 43, 73–81, doi: 10.1016/j.gde.2016.12.007 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Sorrells TR, Booth LN, Tuch BB & Johnson AD Intersecting transcription networks constrain gene regulatory evolution. Nature 523, 361–365, doi: 10.1038/nature14613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beyhan S, Gutierrez M, Voorhies M & Sil A A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol 11, e1001614, doi: 10.1371/journal.pbio.1001614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borneman AR et al. Divergence of transcription factor binding sites across related yeast species. Science 317, 815–819 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Nobile CJ et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148, 126–138, doi:S0092–8674(11)01361–4 [pii] 10.1016/j.cell.2011.10.048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Pastor R, Burchfiel ET & Thiele DJ Regulation of heat shock transcription factors and their roles in physiology and disease. Nature Reviews. Molecular Cell Biology 19, 4–19, doi: 10.1038/nrm.2017.73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernday AD et al. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol 90, 22–35, doi: 10.1111/mmi.12329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB & Johnson AD Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol 5, e256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frazer C et al. Epigenetic cell fate in Candida albicans is controlled by transcription factor condensates acting at super-enhancer-like elements. Nat Microbiol, doi: 10.1038/s41564-020-0760-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chowdhary S, Kainth AS, Pincus D & Gross DS Heat shock factor 1 drives intergenic association of its target gene loci upon heat shock. Cell Reports 26, 18–28 e15, doi: 10.1016/j.celrep.2018.12.034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kainth AS, Chowdhary S, Pincus D & Gross DS Primordial super-enhancers: heat shock-induced chromatin organization in yeast. Trends Cell Biol 31, 801–813, doi: 10.1016/j.tcb.2021.04.004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McSwiggen DT et al. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 8, doi: 10.7554/eLife.47098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blobel GA, Higgs DR, Mitchell JA, Notani D & Young RA Testing the super-enhancer concept. Nat Rev Genet 22, 749–755, doi: 10.1038/s41576-021-00398-w (2021). [DOI] [PubMed] [Google Scholar]
- 87.Chong S et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol Cell, doi: 10.1016/j.molcel.2022.04.007 (2022). [DOI] [PubMed] [Google Scholar]
- 88.Boehning M et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nature Structural & Molecular Biology 25, 833–840, doi: 10.1038/s41594-018-0112-y (2018). [DOI] [PubMed] [Google Scholar]
- 89.Lu H et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323, doi: 10.1038/s41586-018-0174-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burke KA, Janke AM, Rhine CL & Fawzi NL Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell 60, 231–241, doi: 10.1016/j.molcel.2015.09.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo YE et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548, doi: 10.1038/s41586-019-1464-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Portz B & Shorter J Switching condensates: The CTD code goes liquid. Trends Biochem Sci 45, 1–3, doi: 10.1016/j.tibs.2019.10.009 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Quintero-Cadena P, Lenstra TL & Sternberg PW RNA Pol II length and disorder enable cooperative scaling of transcriptional bursting. Mol Cell 79, 207–220 e208, doi: 10.1016/j.molcel.2020.05.030 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Henninger JE et al. RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225 e224, doi: 10.1016/j.cell.2020.11.030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shao W et al. Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nature Chemical Biology 18, 70–80, doi: 10.1038/s41589-021-00904-5 (2022). [DOI] [PubMed] [Google Scholar]
- 96.Bi X et al. RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol Cell 75, 102–116 e109, doi: 10.1016/j.molcel.2019.05.007 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Chen G et al. Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation. Nature Communications 11, 4206, doi: 10.1038/s41467-020-18021-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rencus-Lazar S, DeRowe Y, Adsi H, Gazit E & Laor D Yeast models for the study of amyloid-associated disorders and development of future therapy. Front Mol Biosci 6, 15, doi: 10.3389/fmolb.2019.00015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tuite MF Yeast models of neurodegenerative diseases. Prog Mol Biol Transl Sci 168, 351–379, doi: 10.1016/bs.pmbts.2019.07.001 (2019). [DOI] [PubMed] [Google Scholar]
- 100.Zbinden A, Perez-Berlanga M, De Rossi P & Polymenidou M Phase separation and neurodegenerative diseases: A disturbance in the force. Dev Cell 55, 45–68, doi: 10.1016/j.devcel.2020.09.014 (2020). [DOI] [PubMed] [Google Scholar]
- 101.Darling AL & Shorter J Combating deleterious phase transitions in neurodegenerative disease. Biochim Biophys Acta Mol Cell Res 1868, 118984, doi: 10.1016/j.bbamcr.2021.118984 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun Z et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9, e1000614, doi: 10.1371/journal.pbio.1000614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elden AC et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075, doi: 10.1038/nature09320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shorter J Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev 44, 1–8, doi: 10.1016/j.gde.2017.01.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tariq A et al. Mining disaggregase sequence space to safely counter TDP-43, FUS, and alpha-synuclein proteotoxicity. Cell Reports 28, 2080–2095 e2086, doi: 10.1016/j.celrep.2019.07.069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oldfield CJ & Dunker AK Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83, 553–584, doi: 10.1146/annurev-biochem-072711-164947 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Martin EW et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699, doi: 10.1126/science.aaw8653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pak CW et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell 63, 72–85, doi: 10.1016/j.molcel.2016.05.042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bremer A et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat Chem 14, 196–207, doi: 10.1038/s41557-021-00840-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699 e616, doi: 10.1016/j.cell.2018.06.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vernon RM et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, doi: 10.7554/eLife.31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin Y, Currie SL & Rosen MK Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem 292, 19110–19120, doi: 10.1074/jbc.M117.800466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murthy AC et al. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nature Structural & Molecular Biology 26, 637–648, doi: 10.1038/s41594-019-0250-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Conicella AE et al. TDP-43 alpha-helical structure tunes liquid-liquid phase separation and function. Proc Natl Acad Sci U S A 117, 5883–5894, doi: 10.1073/pnas.1912055117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conicella AE, Zerze GH, Mittal J & Fawzi NL ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549, doi: 10.1016/j.str.2016.07.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Murray DT et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627 e616, doi: 10.1016/j.cell.2017.08.048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kato M et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767, doi: 10.1016/j.cell.2012.04.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kato M, Zhou X & McKnight SL How do protein domains of low sequence complexity work? RNA 28, 3–15, doi: 10.1261/rna.078990.121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fawzi NL, Parekh SH & Mittal J Biophysical studies of phase separation integrating experimental and computational methods. Current Opinion in Structural Biology 70, 78–86, doi: 10.1016/j.sbi.2021.04.004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nature Reviews. Molecular Cell Biology 18, 285–298, doi: 10.1038/nrm.2017.7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Banani SF et al. Compositional control of phase-separated cellular bodies. Cell 166, 651–663, doi: 10.1016/j.cell.2016.06.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mittag T & Pappu RV A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol Cell 82, 2201–2214, doi: 10.1016/j.molcel.2022.05.018 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brangwynne CP Phase transitions and size scaling of membrane-less organelles. J Cell Biol 203, 875–881, doi: 10.1083/jcb.201308087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dutagaci B et al. Charge-driven condensation of RNA and proteins suggests broad role of phase separation in cytoplasmic environments. eLife 10, doi: 10.7554/eLife.64004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dignon GL, Best RB & Mittal J Biomolecular phase separation: From molecular driving forces to macroscopic properties. Annu Rev Phys Chem 71, 53–75, doi: 10.1146/annurev-physchem-071819-113553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garcia-Jove Navarro M et al. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nature Communications 10, 3230, doi: 10.1038/s41467-019-11241-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guillen-Boixet J et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361 e317, doi: 10.1016/j.cell.2020.03.049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maharana S et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921, doi: 10.1126/science.aar7366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ma W, Zheng G, Xie W & Mayr C In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. eLife 10, doi: 10.7554/eLife.64252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


