Figure 1: Motion transmission upon in-silico deactivation of voltage-sensing helix DIII-S4 in the cockroach sodium channel BgNav1–1a.
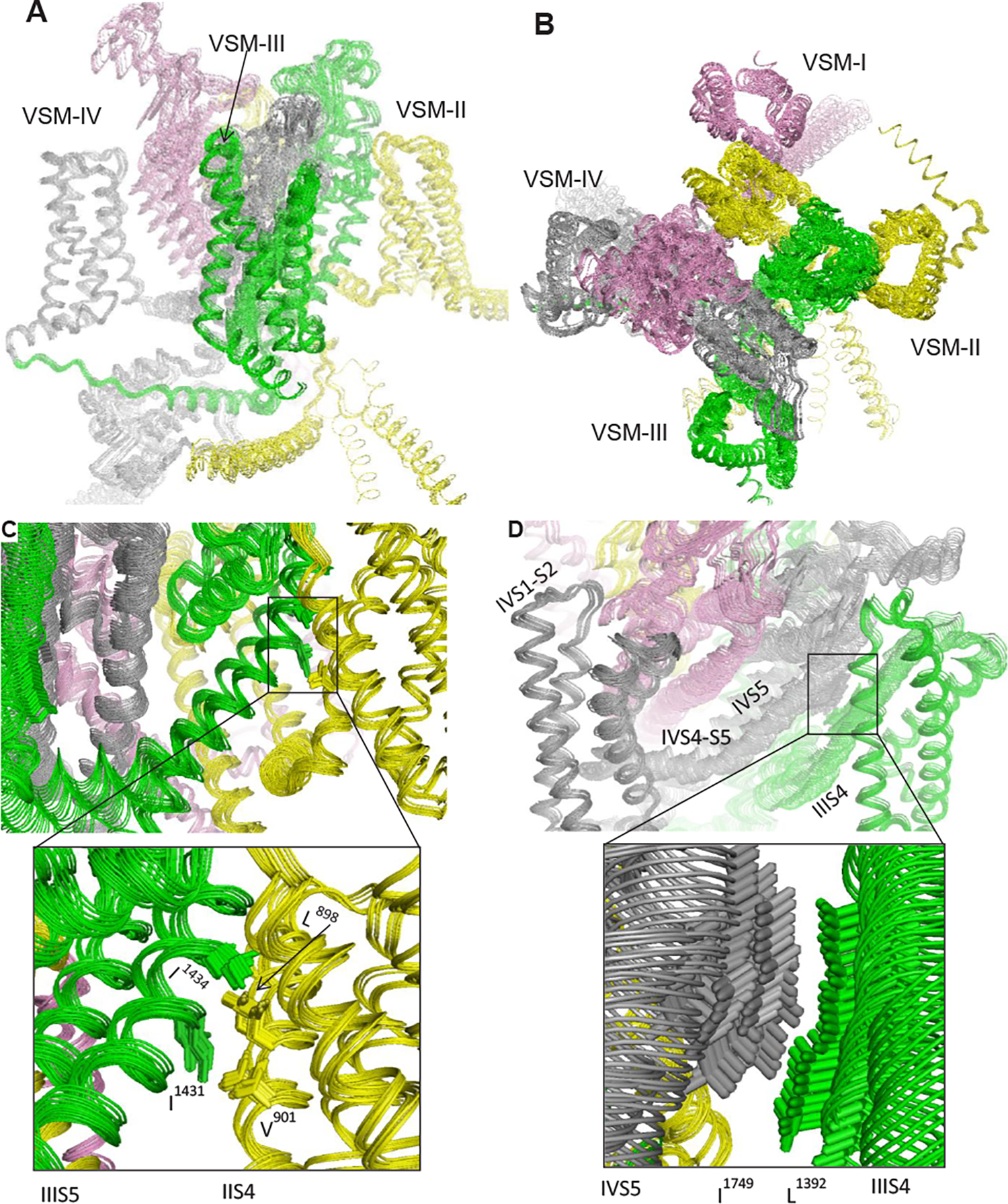
In the intermembrane (A) and extracellular (B) views, repeat domains I, II, III and IV are in pink, yellow, green and gray, respectively. The channel model with activated VSMs and presumably inactivated Pore module was computed at the Compute Canada (www.computecanada.ca) using the AlphaFold2 software [34]. Then the Pore module was in-silico opened towards the cryo-EM structure of the open rNav1.5 channel [35] and helix DIII-S4 was stepwise shifted in the cytoplasmic direction using described methods [29,36]. Twenty-one snapshots were superimposed with PyMol (Version 0.99rc6; Schrödinger, LLC, New York, NY). The forced downshift of helix DIII-S4 caused perturbations all over the channel. In particular, the shift was transmitted to the linker-helix DIII-S4-S5 and then to DII-VSM through hydrophobic contacts between DIII-S5 and DII-S4 (C). Helix DIV-S5 shifted down due to a hydrophobic contact with DIII-S4 (D), and through linker-helix DIV-S4-S5 the motion was transmitted to DIV-VSM.
