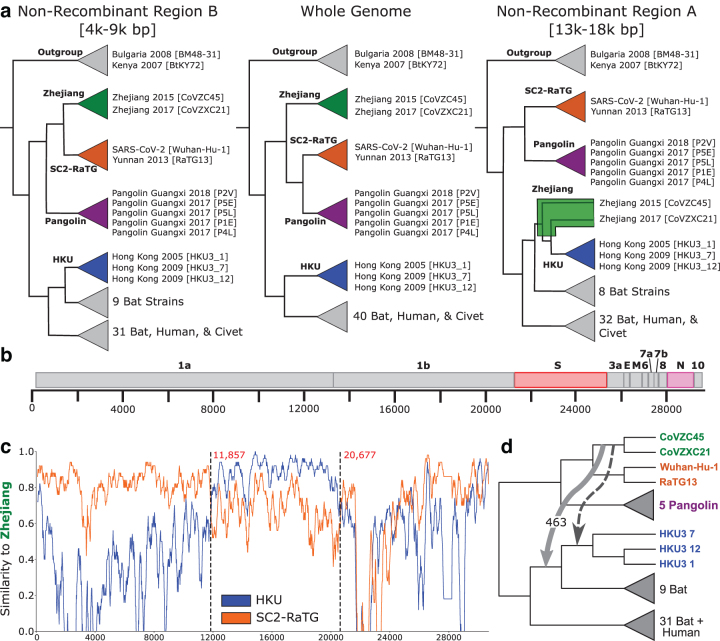
FIG. 2.
Overview of Sarbecovirus genome evolution. (a) We reconstructed three candidate strain trees from the whole genome and two putative nonrecombinant regions A (13,000–18,000 base pairs) and B (4000–9000 base pairs). Their topologies differ substantially, especially in the SARS-CoV-2 lineage, which suggests that the evolution of SARS-CoV-2 was impacted by recombination. We define four clades, Zhejiang (green), SC2-RaTG (orange), Pangolin (purple), and HKU (blue), and show the tree inferred using each region of the genome. (b) The Sarbecovirus genome comprises four well-characterized structural genes which construct the viral spike, envelope, membrane, and nucleocapsid proteins, as well as several open reading frames which encode accessory factors. The spike and nucleocapsid genes are highlighted in red and pink, respectively, as they appear in several ancestral recombinations (Fig. 5). (c) Sequence similarity along the genome using SimPlot. Using Zhejiang clade sequences as query, we compare with the SC2-RaTG and HKU clades. For the majority of the genome, SC2-RaTG is more similar to Zhejiang. Between 11,857 and 20,677 base pairs, HKU is more similar. (d) We find evidence of an HGT from the immediate ancestor of the Zhejiang clade to an ancestor of the HKU clade in ORF1ab. This recombination (light gray) explains the signal shown in the NRR-A tree (a) and SimPlot (c) and is not consistent with the dating of the phylogeny (Supplementary Fig. S2). However, it is not uncommon for inferred HGTs to be off by a single branch due to inference uncertainty. A time-consistent HGT to the ancestor of the three HKU strains (darker gray) similarly explains the signal.