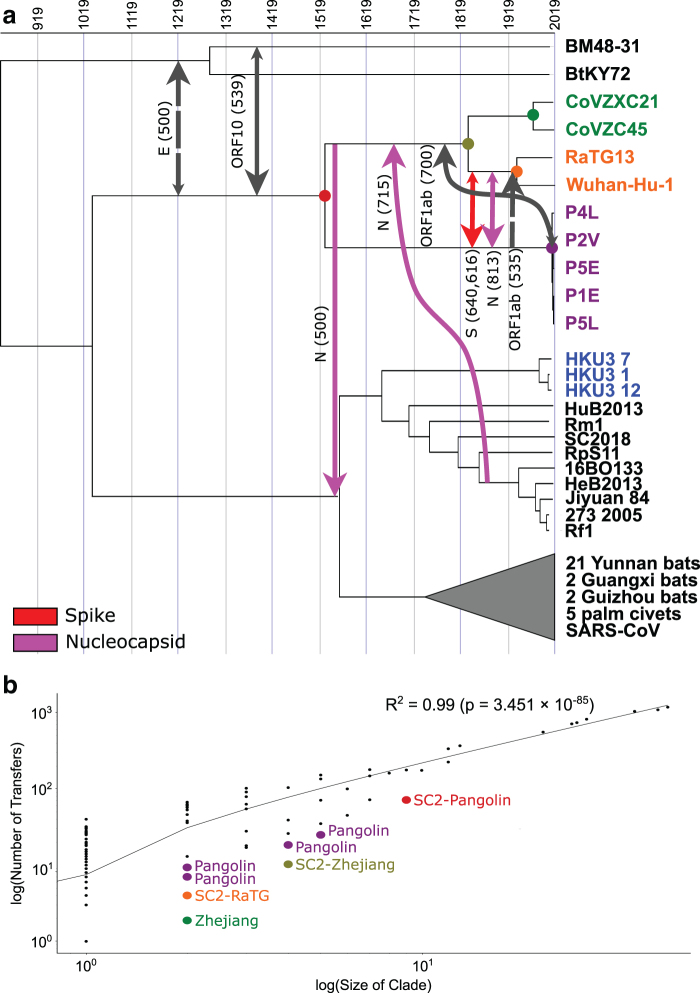
FIG. 3.
HGTs involving the SARS-CoV-2 lineage. (a) We inferred 8 highly supported (with a support of at least 500) HGTs which involve an ancestor of SARS-CoV-2 [Wuhan-Hu-1]. Support values are shown for the OptRoot-rooted gene trees (solid lines) or MAD-rooted gene trees (dashed lines), with one transfer (spike) inferred using both rootings. Smaller arrow heads indicate that there exists an HGT with at least 100 support in the reverse direction using gene trees rooted with either method, suggesting directional uncertainty. (b) We found a strong correlation between the number of leaves in a clade and the number of HGTs identified in that clade (Pearson's R2 = 0.99). However, for every ancestral strain in the SARS-CoV-2 lineage and related clades (highlighted by larger colored points), the number of HGTs in that clade is much lower than would be expected for their size. This paucity of HGTs is likely due to sampling effects, as these strains are more distantly related to the rest of the Sarbecovirus strains in the analysis. MAD, Minimum Ancestor Deviation.