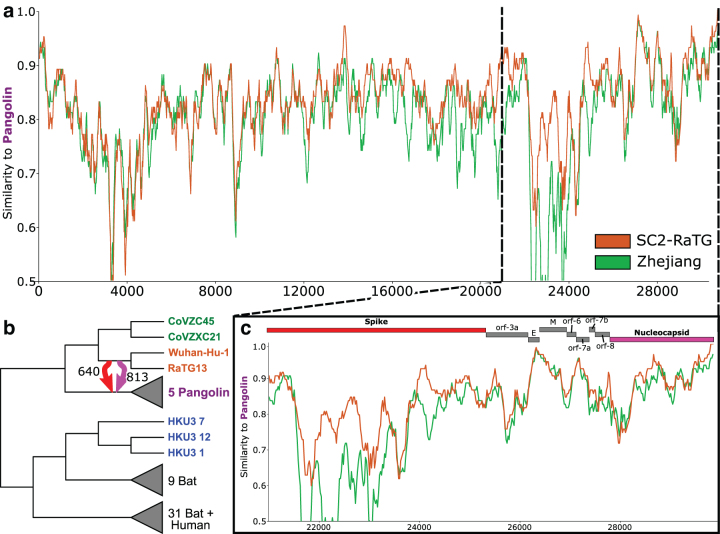
FIG. 5.
Ancestral HGTs are consistent with sequence similarity but difficult to discover from direct sequence comparison alone. We present a case study of an ancestral recombination which is highly supported in both the spike and nucleocapsid gene families, from the immediate ancestor of the SC2-RaTG clade (orange) to the immediate ancestor of the Pangolin clade (purple). (a) For much of the genome, Zhejiang is more similar to the donor SC2-RaTG than the recipient Pangolin. (b) Our analysis infers HGTs from SC2-RaTG to Pangolin in the spike and nucleocapsid genes with supports of 640 and 813, respectively. (c) In the 3′ region of the genome, Pangolin is often more similar to SC2-RaTG, especially in the spike and parts of the nucleocapsid gene families. However, it is difficult to clearly determine from direct sequence comparison alone which gene families have been affected by recombination, especially in ancestral cases such as these where the closest reference relative is the same for both donor and recipient. For this analysis, sequences for ancestral strains were estimated through a majority consensus of their descendants.