Abstract
The specialized cell types of the mucociliary epithelium (MCE) lining the respiratory tract enable continuous airway clearing, with its defects leading to chronic respiratory diseases. The molecular mechanisms driving cell fate acquisition and temporal specialization during mucociliary epithelial development remain largely unknown. Here, we profile the developing Xenopus MCE from pluripotent to mature stages by single-cell transcriptomics, identifying multipotent early epithelial progenitors that execute multilineage cues before specializing into late-stage ionocytes and goblet and basal cells. Combining in silico lineage inference, in situ hybridization, and single-cell multiplexed RNA imaging, we capture the initial bifurcation into early epithelial and multiciliated progenitors and chart cell type emergence and fate progression into specialized cell types. Comparative analysis of nine airway atlases reveals an evolutionary conserved transcriptional module in ciliated cells, whereas secretory and basal types execute distinct function-specific programs across vertebrates. We uncover a continuous nonhierarchical model of MCE development alongside a data resource for understanding respiratory biology.
Single-cell analysis of mucociliary epithelium captures a cell-type and differentiation model, with insights for airway biology.
INTRODUCTION
Embryonic development is a tightly regulated process, where spatiotemporal organizational cues drive cell fate specification and the emerging complexity of cell types within an embryo. The series of single-cell responses and collective cell state transitions guide the organism’s body plan and tissue organization. Of fundamental importance is the development of the vertebrate airway and the lining of the mucociliary epithelium (MCE), which enables fundamental respiratory air exchange and critical defense against inhaled agents, microorganisms, and harmful substances (1–3). During development, the mammalian MCE forms as a pseudostratified complex tissue consisting of multiple cell types, further regulated by chemical and mechanical cues within the epithelium (4, 5). The outer epithelial mature goblet cells secrete mucus that, along with anti-inflammatory peptides, maintains a physical and chemical barrier between the respiratory epithelium and the inhaled air (6). The multiciliated cells and coordinated beating of their cilia propels mucus to the oropharynx where it is expectorated or swallowed (7). The basal cells located below the superficial epithelial cell layer are thought to differentiate into other specialized mucociliary cells across late stages (8), and the proportionally fewer ionocytes control mucus viscosity by balancing electrolyte homeostasis across the epithelium (9). These mature MCE cell types structurally form the organism’s essential respiratory architecture, acting in concert as the primary innate airway defense barrier, and, through efficient mucus clearance, govern essential airway conductance and molecular transport. The increased susceptibility to airway infections, respiratory diseases, and impaired lung function during asthma; chronic obstructive pulmonary disease; and cystic fibrosis is characterized by altered composition, abundance, and distribution of airway MCE cell types (9–12)
The embryonic epidermis of the amphibian Xenopus has emerged as a powerful model to study vertebrate MCE. Similar to the mammalian airway, the Xenopus epidermis develops as a mix of multiciliated and secretory cells (13). The Xenopus and mammalian mucociliary epithelia share notable similarities, with many protein counterparts recently found in mammalian airways (14–17). Recent studies have explored individual MCE cell types and signaling pathways during MCE regeneration and homeostasis across species (9, 11, 12, 18). Single-cell transcriptomics [single-cell RNA sequencing (scRNA-seq)] has been used to profile Xenopus species across embryonic, larval, and adult stages, providing large data resources to study insights into development, regeneration, and repair (19, 20). However, the molecular mechanisms underlying cell fate acquisition, cell type compositions, and temporal dynamics, particularly during MCE development, have been lacking. The tissue from the animal pole of blastula-stage Xenopus embryos (animal cap) composed of pluripotent cell sheets can be cultured as ex vivo explants (organoids) and undergo default mucociliary fate specification into a functional MCE, robustly mimicking the earliest developmental events (21, 22). Here, we profile the entire MCE development across 10 stages through single-cell transcriptomics using Xenopus organoids and perform validation using in situ hybridization and multiplexed spatial RNA imaging in Xenopus embryos. Profiling 33,990 single cells, we characterize the developmental diversification of transitory early-stage cell states and their specialization into basal cells and mature multiciliated, goblet cells and ionocytes. Notably, we capture a multipotent “early epithelial progenitor” population at neurula developmental stages. Applying a generalizable in silico lineage inference method, we demonstrate that basal and secretory (i.e., goblet cell and ionocyte) differentiation requires passage through the multipotent early epithelial progenitors, while ciliated progenitors mature across a distinct lineage into multiciliated cells. We validate the timing, appearance, spatial positioning, and specialization of different cell types through single-cell fluorescence in situ hybridization chain reaction (in situ HCR) and multiplexed RNA imaging. Through comparative analysis of nine cross-species single-cell airway atlases and corresponding cell types, we identify a conserved species-independent expression program in ciliated cells, alongside cell type and species-specific functional programs within basal and secretory cell types (Fig. 1A). Collectively, our work provides a temporal atlas and associated dynamics of emerging specialized cell types, laying the building foundation for dissecting the stepwise formation of complex MCE.
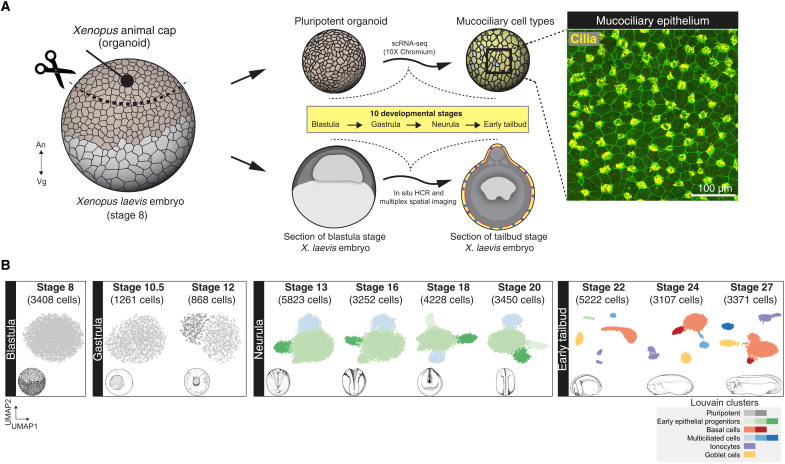
Fig. 1. Cellular composition of developing Xenopus MCE.
(A) Schema for single-cell transcriptome profiling of developing Xenopus MCE. The animal pole tissues from blastula stage 8 embryos were cultured as organoids, sampled temporally across the 10 developmental stages (spanning blastula, gastrula, neurula, and early tailbud stages), and profiled by droplet scRNA-seq. Inset: Surface MCE of stage 27 organoids stained with anti–acetylated tubulin antibody (marking axonemes that build cilia; yellow) and phalloidin (marking filamentous actin; green). (B) Low-dimensional [UMAP (uniform manifold approximation and projection)] scRNA-seq visualization of the different developmental stages and Louvain clusters, colored based on major cell types.
RESULTS
Single-cell transcriptomic profiling of developing MCE
To generate a continuous fate map of developing Xenopus MCE, we profiled animal cap organoids corresponding to 10 developmental stages from early pluripotency to a functional MCE spanning blastula [Nieuwkoop and Faber (NF) stage 8], gastrula (stages 10.5 and 12), neurula (stages 13, 16, 18, and 21), and early tailbud development (stages 22, 24, and 27) (Fig. 1A and Materials and Methods) (23), obtaining transcriptomes of 33,990 single cells (fig. S1, A to C; Materials and Methods; and note S1). The initial assessment of individual stages by unsupervised clustering revealed the formation of all known major MCE cell types at early tailbud stages and across several subclusters (Fig. 1B; fig. S2, A to C; and table S2) (24, 25).
At the blastula stage, the fate-unspecified pluripotent cells formed a homogeneous population expressing pluripotency transcription factor pou5f3 and other markers of the undifferentiated state such as sox3 (Fig. 1B and fig. S2A) (26, 27) and could be further distinguished into two clusters by gastrula stages (dark and light gray clusters; Fig. 1B and fig. S2A). The neurula stages (stages 13 to 20) were characterized by a complete loss of pluripotency and emergence of previously unknown progenitor populations (green and blue clusters; Fig. 1B and fig. S2B). We termed the larger population as early epithelial progenitors (three subclusters), expressing stochastic, low-expression levels of multiple genes such as extracellular matrix component (has1) and elongation factor protein (eef1a1o) at single-cell level (Fig. 1B and fig. S2B). By late neurula, the individual early epithelial progenitor subclusters expressed secretory markers [goblet: glycoprotein otogelin (otog; also known as mucXS and otogl2) and ionocytes: vacuolar-type adenosine triphosphatase (ATPase) proton transporter (atp6v1g3)], indicating their maturation into goblet cells and ionocytes (fig. S2B). The smaller neurula-stage population (light blue; fig. S2B) expressed microtubule components (β-tubulin: tubb4b; dynein: dynll1), indicating a bias toward multiciliated cells. The tailbud stages (stages 22 to 27) marked the respective specialization of the early multiciliated population into multiciliated clusters and early epithelial progenitor subpopulations into ionocytes, goblet, and basal cell clusters (Fig. 1B and fig. S2C). The initial stage-specific scRNA-seq analysis captured major MCE cell types but with previously unknown heterogeneity within cell types.
Integrated time course analysis of mucociliary epithelial development
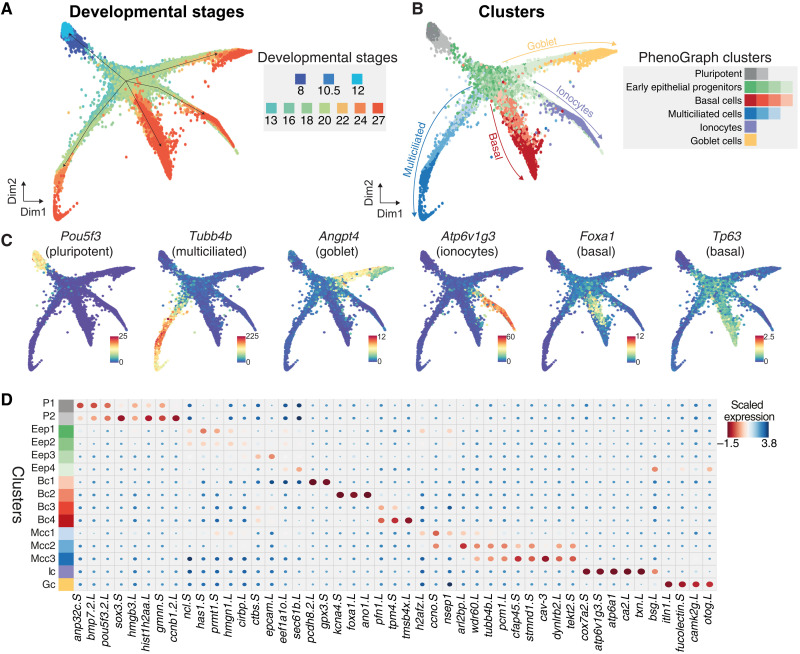
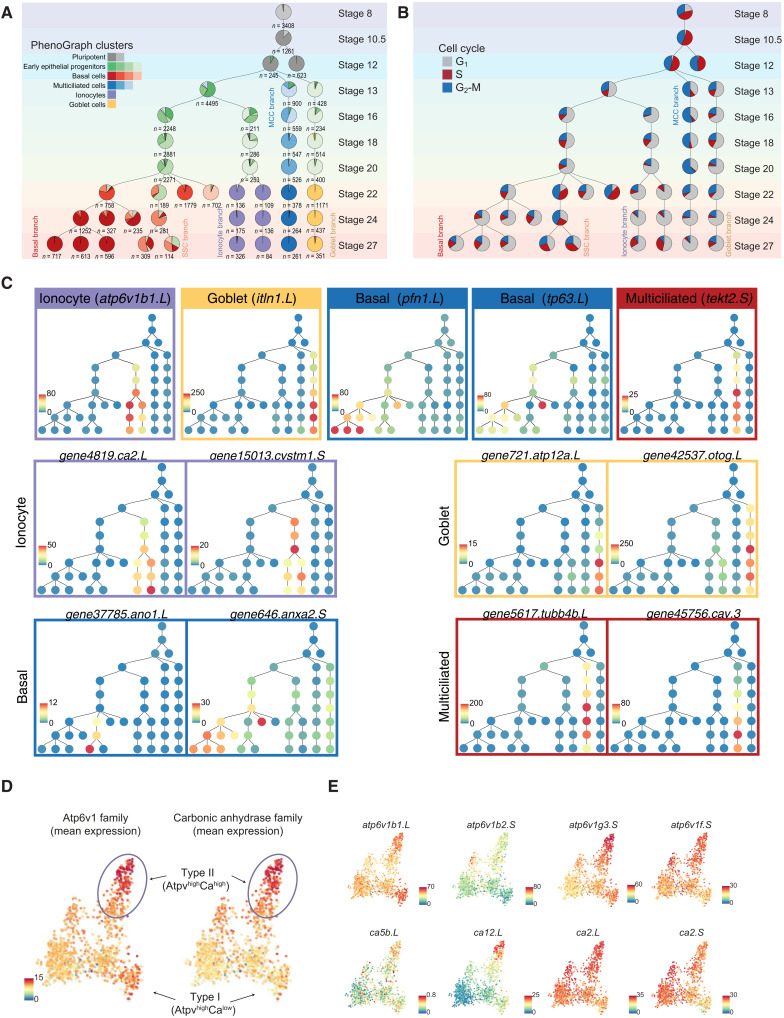
To capture the developmental progression and single-cell state transitions over the entire MCE development, we integrated, visualized, and analyzed all the stages together over a graph embedding of the developmental manifold (Fig. 2A and table S5). This low-dimensional embedding mimicked a starfish pattern, with the blastula and gastrula stages at the head (stages 8, 10.5, and 12) separated from the late stages across individual arms (stages 22 to 27). We observed that graph-based approaches were suited for visualizing such high-density time series datasets, and community-based clustering approaches failed to resolve meaningful cell states and types (fig. S3D). To classify cell states over the continuous high–cell density MCE developmental manifold, we adapted and applied PhenoGraph clustering (28) and identified five major cell types, further separated into 15 different clusters (Fig. 2B; fig. S3, A and B; and tables S3 to S6). These clusters included two pluripotent (P1 and P2), four early epithelial progenitors (Eep1, Eep2, Eep3, and Eep4), four basal (Bc1, Bc2, Bc3, and Bc4), three multiciliated (Mcc1, Mcc2, and Mcc3), an ionocyte (Ic), and a goblet cell cluster (Gc) (Fig. 2, A and B, and fig. S3, A and B). The clusters were named based on the expression of known and additional cell type–specific markers (Fig. 2, C and D, and figs. S3C and S4, A to F). Notably, the cluster membership spanned single cells from multiple developmental stages and scRNA-seq runs, indicating that clusters are biologically relevant and not because of technical or batch effects (Fig. 2, A to D, and figs. S3C and S4, A to F). While the major cell types are discrete, the cell states (i.e., subclusters) are transitory and describe the different trajectories taken by single cells over the developmental MCE manifold.
Fig. 2. Joint embedding of continuous MCE developmental manifold and cell-type differentiation.
(A) High-density force-directed k-nearest neighbor (knn) graph visualization of single cells, colored by developmental stages. (B) PhenoGraph clustering of single cells over continuous MCE manifold, colored by different cell types and cell states (subclusters). The arrows indicate the differentiation of progenitors into specific cell types. (C) Expression patterns of marker genes for MCE cell types overlaid on the knn graph. The color bars indicated scaled imputed expression levels. (D) Dot plot of marker genes across different MCE clusters. The color represents maximum normalized mean marker gene expression across each cluster, and the size indicates the proportion of positive cells relative to the entire dataset.
The pluripotent clusters (P1 and P2) expressed the respective transcription factors (TFs) (pou5f3 and sox3), mRNA splicing factors (srsf5 and srsf7), signaling molecules (bmp7.2), and cell cycle regulators (anp32c and gmnn), indicating a hypertranscriptional activity and fast cycling cells (Fig. 2C and figs. S3C and S4A), consistent with the highest number of highly variable genes (HVGs) (fig. S1C). The P2 cluster could be distinguished from the P1 cluster based on the higher expression of krt5.7.L, upk3b.L, and grhl3.S, which become restricted to cells present in the outer or superficial layers (29). The early epithelial progenitors (Eep1 to Eep4) expressed several general TFs (nsep1, prmt1, and hmgn1) and genes with broad cellular functions including adhesion (epcam), poly-A binding (ncl and cirbp), and ribosomal genes, indicating the lack of a specific transcriptional program (figs. S3C and S4B). The basal cells across clusters (Bc1 to Bc4) expressed increasing levels of forkhead box TF (foxa1), ectoderm-specifying TF (tp63), actin-binding proteins (pfn1 and tmsb4x), and ion channel members (ano1 and kcna) (Fig. 2C and figs. S3C and S4C). The multiciliated cells across clusters (Mcc1 to Mcc3) express increasing levels of structural (tekt2), cilia (cfap45), and cytoskeletal components (dynlrb2 and tubb4b) (Fig. 2C and figs. S3C and S4D) as well as caveolin (cav3) that regulates cilia length and beat frequency (2, 30). The joint embedding and clustering identified an ionocyte and goblet cell cluster, respectively. The osmoregulatory, proton-pumping ionocytes (Ic) expressed forkhead transcription factor I1 (foxi1), multimeric V-type ATPase family (v1a, v1g, and v0d), cytochrome oxidase subunits (cox7a2), and carbonic anhydrases (ca12), catalyzing protons and bicarbonate transfer (Fig. 2C and figs. S3C and S4E), while the goblet cells (Gc) expressed angiopoietin (angpt4), lectins (itln1 and fucolectin), and unconventional mucin glycoprotein otogelin (otog; also known as mucXS and otogl2) (Fig. 2C and figs. S3C and S4F).
The high-dimensional integrated analysis not only captured the major MCE cell types based on expected marker gene expression but also revealed additional insights. First, the complete continuous MCE development could be integrated and visualized in a low-dimensional space, capturing the cell state specialization and transitions across branches. The homogeneous pluripotent cells undergo specialization at early neurula, forming highly heterogeneous early epithelial progenitor clusters (Fig. 2, A and B). The broadly dispersed early epithelial progenitor states across the developmental manifold express stochastic levels of multiple markers, retain transcriptional plasticity and competence to respond to multilineage cues (by stages 18 to 20), and execute cell type–specific programs at tailbud stages. Second, we observed multiciliated progenitors by stage 13 (alongside early epithelial progenitors), marking an early priming event for ciliogenesis. Third, by stage 27, the basal cells developed alongside multiciliated, ionocyte, and goblet cells as the most abundant cell type (i.e., the proportion of cells/total sample proportion per stage; Materials and Methods). Notably, the observed basal cell differentiation dynamics were different from other cell types, owing to the presence of multiple late-stage subclusters and the expression of distinct TFs. We observed low levels of TF tp63 within early epithelial progenitors (primed to form basal cells), and tp63 expression increased specifically across basal cell clusters at tailbud stages. These late-stage basal clusters were highly heterogeneous and constituted cells expressing either tp63 alone or both tp63 and foxa1 (Fig. 2C). Last, standard community–based clustering approaches were suboptimal than graph-based methods (applied here) at deconvoluting the continuous MCE manifold, owing to irregular and highly continuous density of single cells over the MCE developmental time course (fig. S3D). Overall, the profiled scRNA-seq MCE developmental atlas and analysis reliably captured the developmental transitions starting from pluripotent stem cells to specialized mucociliary epithelial cell types.
Developmental trajectories and cell type formation during MCE development
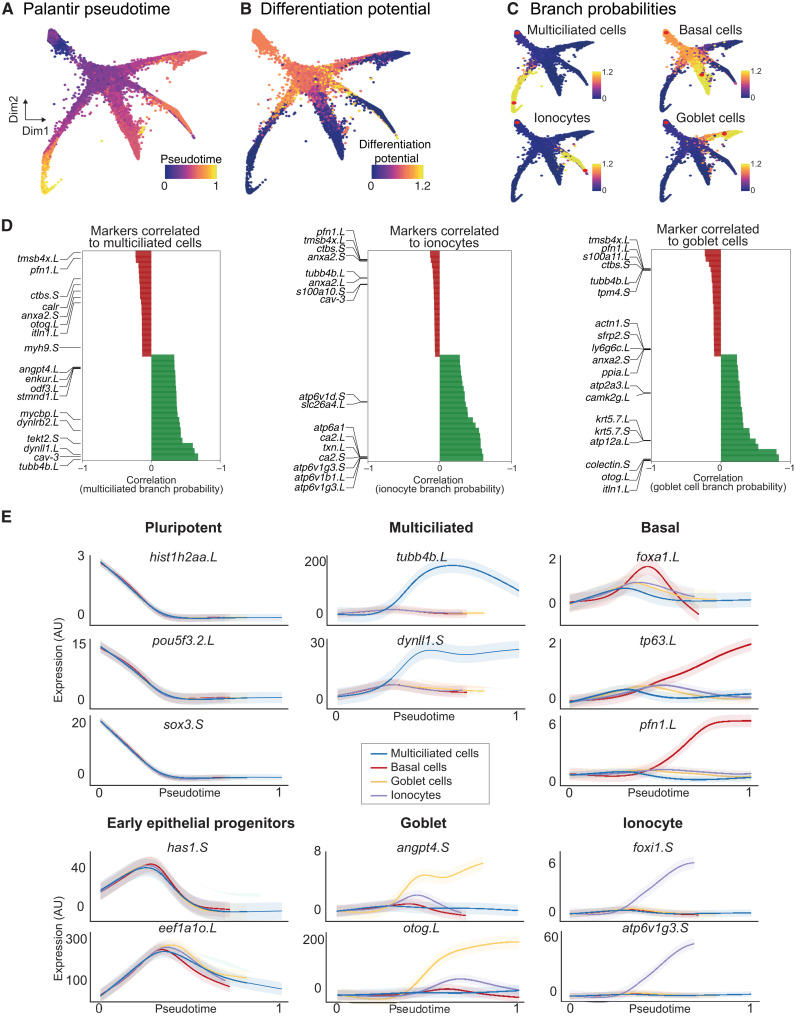
To characterize the developmental trajectories and maturation of different cell types during MCE development, we (i) calculated independent pseudotime ordering of all single cells (Fig. 3A), (ii) computed single-cell differentiation potential (Fig. 3B), and (iii) inferred individual branch probabilities for pluripotent cells to reach different discrete cell types (Fig. 3C).
Fig. 3. Developmental transitions and cell state branching over MCE trajectories.
(A) Pseudotime inferred by Palantir overlaid on the MCE manifold (knn graph). The pluripotent cells mark the beginning of pseudotime, while terminal cell states (Mcc, Gc, Ic, and Bc) mark late pseudotime. (B) Differentiation potential overlaid on the MCE manifold (knn graph) exclusively marks pluripotent and progenitor populations. (C) Branch probabilities assessed for each late-stage cell type and overlaid over on the MCE manifold (knn graph). The red dots mark the start (pluripotent) and respective end cells (cell types). (D) Marker gene correlations to multiciliated, ionocytes, and goblet cell branches of the MCE manifold (knn graph). Green and red colors signify positively and negatively correlated genes, respectively. (E) Gene expression trends of marker genes, using generalized additive models, contributing to individual branch probabilities including pluripotent (hist1h2aa.L, pou5f3.2.L, and sox3.S), early epithelial progenitors (has1.S and eef1ao.L), multiciliated (pink; tubb4b.L and dynll1.S), basal (green; foxa1.L, tp63.L, and pfn1.L), goblet cells (orange; angpt4.S and otog.L), and ionocytes (blue; foxi1.S and atp6v1g3.S). The variable pseudotime reflects distinct cell type diversification and differentiation speeds over MCE development. AU, arbitrary units.
We applied Palantir, to infer an unbiased pseudotime over the continuous high-density developmental manifold, as conventional approaches failed to generate cell type–specific trajectories. The pseudotime is inferred as a probabilistic process based on entropy over the entire manifold, with different maturation dynamics for each cell type (Fig. 3, A and E). As expected, we observed that cells from blastula and neurula stages (pluripotent, early epithelial, and ciliated progenitors) had low pseudotime, which increased across the branches consisting of specialized multiciliated, basal, ionocyte, and goblet cells (Fig. 3A). The multiciliated branch had the highest pseudotime, indicating that they were fully differentiated, relative to other cell types (Fig. 3A).
As an independent measure of cellular plasticity, we calculated differentiation potential per single cell (Materials and Methods) and observed that pluripotent (P1 and P2), ciliated (Mcc1), and early epithelial progenitors (Eep1 to Eep4) had the highest differentiation potential, markedly declining with lineage commitment across four branches (Fig. 3B). The differentiation potential strongly phenocopied the cell state assignment (Fig. 2, A and B) and was inversely correlated with pseudotime (Fig. 3, A and B), validating the cell states (subclusters) and cell type assignment (multiciliated, basal, goblet, and ionocytes) by multiple independent computational approaches.
As additional validation of cellular trajectories, we calculated branch probabilities of single cells through iterative refinement of the shortest distance over the MCE manifold, assuming unidirectional progression and independence from neighboring cells (Fig. 3C). The branch probabilities indicate the likelihoods of a pluripotent cell (red start cell at top) ending up as a differentiated and discrete cell type (red end cell within multiciliated, basal, goblet, and ionocytes; Fig. 3C and Materials and Methods). The branch probabilities were low across pluripotent, early, and multiciliated progenitors and markedly increased across all late-stage cell types, except basal cells (Fig. 3C). The branch probabilities of basal cells had a marginal increase (relative to multiciliated, goblet, and ionocytes; Fig. 3C, top right), indicating that the basal cells share properties with pluripotent cells and act as late-stage stem cell reservoirs (31, 32). We further correlated the expression of known markers within individual branches and identified additional genes driving cell type differentiation within single cells (Fig. 3D). We observed that many genes had nonlinear expression patterns and used generalized additive models to visualize averaged expression over respective cell type pseudotime (Fig. 3E). The pluripotency markers (pou5f3, sox3, and hist1h2aa) were highly expressed in early pseudotime across all cell types and decreased with cell type differentiation. Most cell type–specific markers including multiciliated (tubb4b and dynll1), basal (tp63 and pfn1), goblet (angpt4 and otog), and ionocytes (foxi1 and atp6v1g3) showed nonlinear trends, selectively peaking at respective trajectories over the MCE pseudotime (Fig. 3E). Consistent with multipotent roles, the early epithelial progenitors peaked across all trajectories but sharply declined before specialization (Fig. 3E). Notably, generalized additive models highlight that single-cell progression through the individual pseudotime lacked well-defined bifurcation points, further validating that early epithelial progenitors are plastic and undertake continuous cell fate choices. Unexpectedly, the expression dynamics of basal markers (foxa1, pfn1, and tp63) strongly differed across pseudotime, highlighting heterogeneity and differential regulation of subclusters (Bc1 to Bc4) and spatial positioning within sensorial layers (Figs. 3E and 8B). The foxa1 peaked in the middle (restricted to basal subcluster), while tp63 and pfn1 linearly increased over all basal cell pseudotime, distinct from other cell types over MCE pseudotime (Fig. 3E). The generalized additive models confirm the robustness of the trajectory analysis (pseudotime, differentiation potential, and branch probabilities) and further highlight that basal cell differentiation dynamics is different to other MCE cell types.
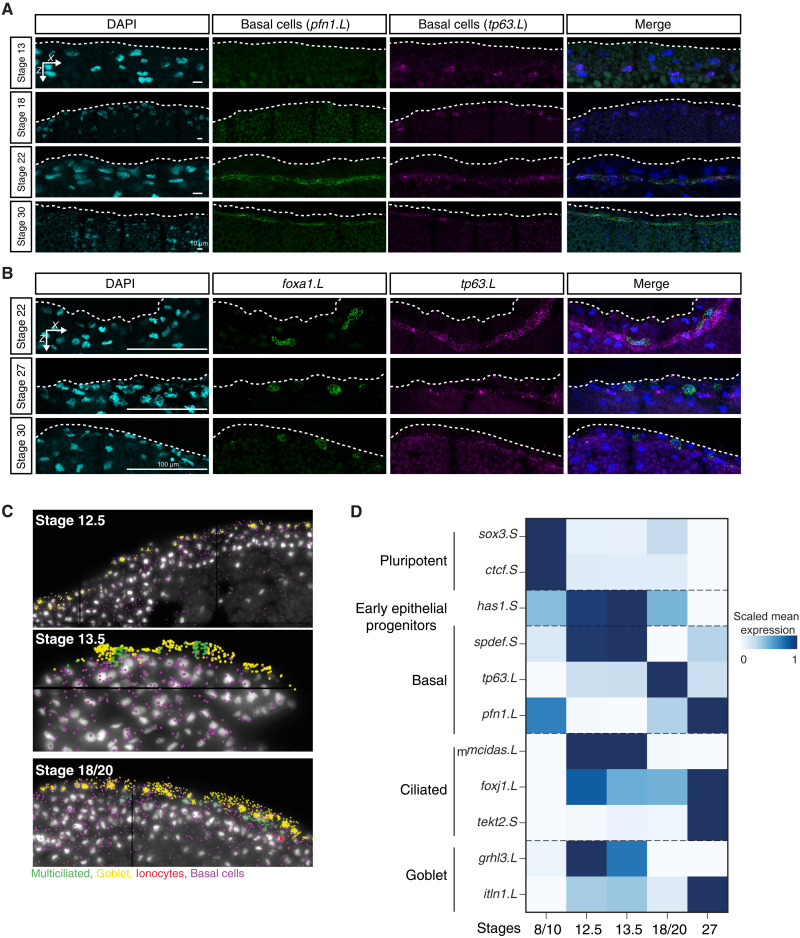
Fig. 8. In situ HCR validation of developing basal cell lineage over MCE development.
(A) In situ HCR of apical superficial epithelium indicating basal cells marked by profilin (pfn1.L; green), tumor protein P63 (tp63.L; magenta) across neurula (stages 13 and 18), and tailbud stages (stages 22 and 30); nuclei are marked by DAPI staining (in cyan, separate channel and in blue, merged channels). The expression levels of pfn1.L and tp63.L decrease by stage 30. Images (here and below) represent the maximum intensity projections of transverse cross sections, and white dashed lines indicate the apical superficial epithelium. (B) In situ HCR marked by forkhead box A1 (foxa1.L; green) and tumor protein P63 (tp63.L; magenta) across tailbud stages. (C) Representative images of single-cell multiplexed RNA imaging in whole embryos across three MCE developmental stages. Stages 12.5 and 13.5 mark the transition into neurula development. (D) Heatmap showing the RNA abundances of different cell type markers, as captured by multiplexed single-cell RNA imaging. The early epithelial progenitors are marked by the expression of multiple markers at early stages, while specialized cell types exclusively express markers at late stages. The color bars indicate z-scaled mean expression in aggregated cells.
Next, we computed single-cell RNA velocities as a predictive model of differentiation dynamics, i.e., direction and speed of a single cell across complex developmental trajectories, by inferring a per-gene reaction model relating to abundance of unspliced and spliced mRNA (33, 34). The velocity estimates confirmed homogeneity in blastula and gastrula stages and progression through progenitors to the discrete cell types at tailbud stages (i.e., nonoverlapping arrows within discrete clusters). The early epithelial progenitors across neurula stages (stages 13 to 20) were highly plastic (smaller arrows) and separated from multiciliated progenitors (Mcc1 to Mcc3; fig. S5A).
We next focused on the characteristics of individual cell types (figs. S6 and S7). Zooming in on the early epithelial progenitors, we observed that single cells within subclusters (Eep1 to Eep4) were not uniquely biased but had shared propensities toward ionocytes (foxi1, ca2, and bsg), goblet (otog, itln, and fucolectin), and basal cells (tmsb4x, has1, and ctbs), confirming early epithelial progenitors as a multipotent population (fig. S6, A and B). Contrary to early epithelial progenitors, the multiciliated cell differentiation followed stepwise progression starting from multiciliated progenitors (Mcc1; stages 13 to 20) and maturation into multiciliated cells (Mcc2 and Mcc3, tailbud stages; fig. S6, C and D). The tailbud-stage basal cell subclusters (Bc1 to Bc4; stages 22 to 27) located within the deep sensorial layers could be distinguished on the basis of the expression of tp63, pfn1, mal2, and foxa1 (fig. S7, A to C). The Bc1 subcluster has the highest proliferating rates (S phase) and is present throughout tailbud stages (similar proportion of cells; fig. S7C). The proliferation rates decrease from Bc1 to Bc4 (decreasing S phase, increasing G1 cells), indicating a reduced cell cycle speed. Both goblet and ionocytes underwent temporal specialization from stages 22 to 24 (fig. S7, D and E). We observed two separate ionocyte maturation trajectories across tailbud stages (fig. S7E), which we further distinguish as type I (alpha ionocytes; higher V-type ATPase and lower carbonic anhydrase expression) from type II (beta ionocytes; higher V-type ATPase and carbonic anhydrase expression) ionocytes (Fig. 5, D and E, and fig. S12A) and as previously observed in Xenopus and mammalian kidney (35, 36). In summary, we capture the nonhierarchical mode of MCE lineage commitment that starts with the emergence of multipotent early epithelial progenitors that stochastically execute transcription programs within a defined developmental window (neurula stage), followed by maturation into different specialized cell types (basal, ionocytes, and goblet cells) at a similar developmental time, but with different dynamics and maturation speeds.
Fig. 5. Lineage inference of developing MCE cell types.
(A) In silico lineage inference of the cell states and types across developing MCE. The number of cells within each group are highlighted underneath each node. The pie chart indicates proportions of cells mapping to clusters (consensus voting; see Materials and Methods). (B) Lineage inference map overlaid with proportions of cells across cell cycle stages. (C) Timing and expression of different marker genes (ionocytes: atp6v1b1.L; goblet: itln1.L; basal: pfn1.L and tp63.L; multiciliated cells: tekt2.S) over the inferred lineage tree. Lineage restricted expression of additional markers of ionocytes (ca2.L and cystm1.S), goblet (atp12a.L and otog.L/mucXS/otogl2.L), basal (ano1.L and anxa2.S), and multiciliated cells (tubb4b.L and cav3.S). (D) Low-dimensional visualization of ionocyte subpopulations, overlaid with the mean expression of Atp6v1 and carbonic anhydrase family members, indicating type I (Atp6v1highCalow) and type II (Atp6v1highCahigh) ionocytes. (E) Low-dimensional visualization of ionocyte subpopulations overlaid with expression of few Atp6v1 and carbonic anhydrase family members. The color bars indicate the scaled imputed expression of respective markers.
Cell type–specific TFs and signaling pathways during MCE development
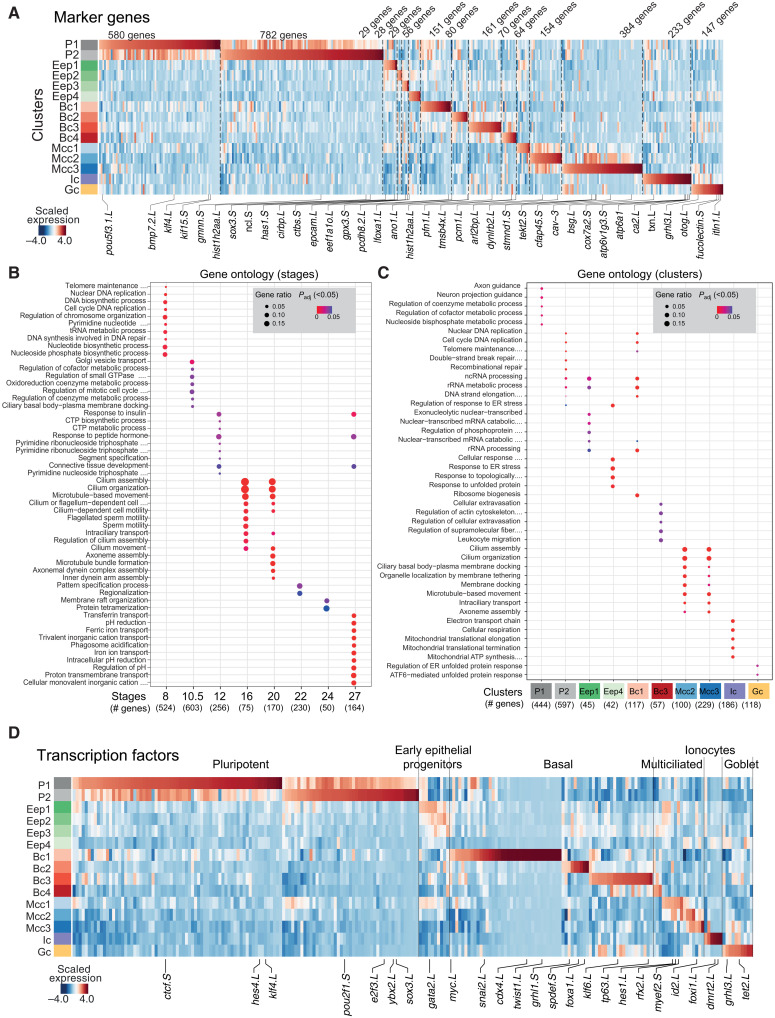
To identify the timing and order of transcriptional programs, we analyzed the expression dynamics of differentially enriched TFs (37) and signaling pathways across MCE development (Materials and Methods and table S6). The pluripotent clusters globally expressed the highest number of genes (Fig. 4, A, D, and E, and fig. S1C), involved in most cellular processes such as metabolic, cell cycle, and repair [gene ontology (GO) terms; Fig. 4, B and C]. The early epithelial progenitors had few/no enriched genes and shared GO terms with other cell types. The multiciliated clusters were enriched with cilia, microtubule-related terms, while the ionocytes were enriched for ion transport and adenosine triphosphate hydrolysis (Fig. 4, A to C). Most TFs were enriched across pluripotent clusters (pluripotent: POU, SOX, yy1, ctcf, and HMG family; remodelers: SMAD, arid2, and jarid family; activators: znf326 and znf706l) and were down-regulated with lineage specification (Fig. 4D and table S6). We identified known and additional cluster-specific TFs for multiciliated (nsep1, ybx1, hes1, rfx2, id2, id4, and sp7), goblet cells (grhl1, grhl3, nkx2-3, atf4, and hic2), and ionocytes (akna, foxi1, dmrt2, tef, and esrra). The basal TFs included developmental [homeobox, caudal ParaHox, and bHLH (basic helix-loop-helix) family], secretory (foxa1, gfi1, tcf25, and pbx2), and growth regulators (tp63, tbx3, tbx2, and usf1).
Fig. 4. Expression of marker genes, TFs, and gene ontology (GO) terms over MCE development.
(A) Heatmap showing differentially expressed markers across MCE clusters. (B) GO (biological processes; P < 0.05) terms for each MCE developmental stage. (C) GO (biological processes; P < 0.05) terms for each PhenoGraph cluster. Five clusters had no significant terms and were not presented. The gene ratio highlights the percentage of total GO term genes identified as enriched within each group (developmental stage/PhenoGraph cluster). (D) Heatmap showing the expression pattern of differentially expressed TFs (x axis) over MCE PhenoGraph clusters (y axis). The color bars (A) and (D) indicate the z-scaled mean expression of respective markers. Padj, adjusted P value.
Although blastula-stage organoids are cultured in the absence of signaling factors and morphogens, several signaling pathways and gradients contribute to their default ex vivo specification into MCE cell types. We observed enrichment of multiple noncanonical signaling pathways across pluripotent [Notch, Wnt, and transforming growth factor–β (TGF-β); fig. S8A and table S7] and basal cell clusters, while the specialized cell types (multiciliated, goblet, and ionocytes) had fewer and highly restricted expression (fig. S8A and table S7). Notably, we observe stage-specific up-regulation of Notch ligands and receptors in basal cell subclusters (notch2, smad3, and smad7) and ciliated progenitors (Mcc1: hes1 and hes8) (fig. S8A and table S7). We next compared lineage-specific TFs and known marker genes identified from bulk transcriptomics (38) and observed a notable single-cell heterogeneity, particularly across multiciliated cells, ionocytes, and basal cells (fig. S8, B to D; tables S6 and S7; and note S2). The reported multiciliated markers at single-cell level had a stepwise activation from Mcc1 (progenitors) to Mcc3 (mature), averaged and missed across bulk assays (fig. S8B), while only a subset of bulk ionocyte markers was strongly enriched at single-cell level across ionocytes (fig. S8C) (18). We observed a similar pattern with recently published basal cell markers (fig. S8D) (18). The expression heterogeneity at single-cell level and discordance with bulk expression indicate that TF activity within subclusters is tightly coupled with cell type maturation, and cell type–specific TFs have a high degree of positive regulation during MCE development.
Intrigued by the expression discordance of bulk markers at single-cell level, we investigated how molecular features (expression noise, coefficient of variation, and transcript diversity) varied over MCE development. Within populations, individual cells have been reported to resolve cellular decisions between stemness and uncertainty through increased heterogeneity and changes across molecular features to drive noise-induced differentiation (37, 38). We observed that both the coefficient of variation and entropy increased over MCE development and was highest at the tailbud stages (fig. S9, A and B). Notably, transcriptional diversity (number of unique transcripts) was anticorrelated and decreased as single cells underwent lineage commitment (fig. S9, A and B), indicating an inherent stochastic gene expression within neurula-stage progenitors contributing to their plasticity through the expression of multilineage expression programs (low expression of many unique transcripts). The progression to late stages leads to the execution of specific cell type gene regulatory networks (increased expression of fewer specific transcripts; higher variance, low transcriptional diversity) (39). We also reanalyzed the MCE atlas through the recent Cellular Trajectory Reconstruction Analysis (CytoTRACE) framework, which measures expressed genes per cell as a proxy for transcriptional diversity and cell type differentiation (40). We confirmed that early and multiciliated progenitor clusters (Eep1 to Eep4 and Mcc1 and Mcc2) had a low CytoTRACE score, indicating multipotent characteristics, while the late-stage cell types (Bc1 to Bc4, Mcc3, Ic, and Gc) had the highest scores, indicating a mature state (fig. S9, C and D), further validating that MCE developmental decisions are modulated through changes in molecular features. The summarized comparison of different molecular features is highlighted in fig. S9E (tables S1 and S9). Together, our analysis suggests that the neurula stages provide the developmental window for individual cells to exhibit different cell fate switching frequencies and establish initial heterogeneities that are resolved by cell type–specific regulatory networks.
In silico cell lineage inference and in situ validation of cell type dynamics across MCE development
Given our time-resolved MCE developmental scRNA-seq atlas, we devised an in silico lineage inference algorithm to map cell state and cell type relationships (Fig. 5, A to D) and validated their temporal emergence and dynamics in whole embryos using in situ HCR and multiplex spatial imaging. While lineage inference from scRNA-seq data is nontrivial, we devised a generalizable method built on a neighborhood mapping and consensus voting strategy, requiring few/no priors to map the single cells and their cluster relationships between successive developmental stages, and charted the MCE development (Fig. 5, A and B; Materials and Methods; and note S3). The inferred lineage tree provided several insights into developmental timing, TFs, marker gene expression, cell cycle, and state relationships over MCE development (Fig. 5, A to C).
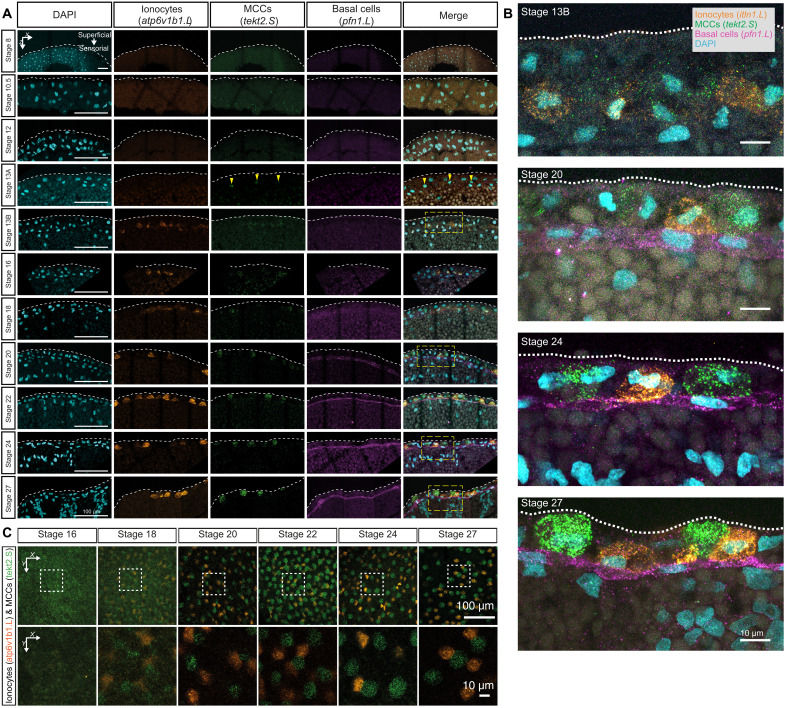
First, the multiciliated transcriptional program was initiated directly from the pluripotent tissue as a distinct lineage (mcidas and tekt2 expression at stage 13) and underwent maturation into multiciliated cells, with decreasing cell cycle speeds and proportionally fewer cells, relative to the total cellular pool (blue branch; Fig. 5, A to C). The single-cell in situ HCR in embryos confirmed the timing and lineage activation of multiciliated progenitors (tekt2) by stage 13, mutually exclusive with other cell types (Fig. 6, A to C, and fig. S10A). We observed a continuous increase of the tekt2 mRNA fluorescent signal in the cytoplasm as multiciliated progenitors radially intercalated (i.e., apical movement) from the deep sensorial layers and integrated within the superficial epithelium at stage 24 (Fig. 6, A to C, and fig. S10A) (41).
Fig. 6. In situ HCR validation of multiple cell type lineages over MCE development.
(A) In situ HCR and validation of lineage inference over 10 development stages of the embryonic epidermis in embryos, marking ionocytes (atp6v1b1.L; orange), multiciliated (tekt2.S; green), and basal cells (pfn1.L; magenta). The nuclei are marked by 4′,6-diamidino-2-phenylindole (DAPI) staining (cyan). The multiciliated cells and ionocytes emerge during neurula in the sensorial layer and radially intercalate (move apically) to superficial layers by early tailbud stages. Multiciliated cells are consistently observed at stage 13 (stage 13A), whereas ionocytes are consistently observed at stage 16; in four of nine embryos, ionocytes are observed at stage 13 (stage 13B). The basal cells are retained in the sensorial layer. The yellow dashed rectangles indicate the zoomed-in regions shown in (B). Yellow arrowheads point to regions (within the nucleus) expressing tekt2.S. (B) Zoomed view of in situ HCR marking expression, positioning, and migration of multiciliated (tekt2.S; green), ionocytes (atp6v1b1.L; orange), and basal cells (pfn1.L; magenta). Images in (A) and (B) represent the maximum intensity projections of transverse cross sections, and white dashed lines indicate the apical superficial epithelium. (C) In situ HCR of whole embryonic surface epithelium marked by multiciliated (tekt2.S; green) and ionocytes (atp6v1b1.L; orange) at the superficial epidermis. Multiciliated cells and ionocytes move apically (radially intercalate) from the sensorial to the superficial epithelium and distribute in a salt-and-pepper fashion. Images represent the maximum intensity projections of Z sections taken from the apical superficial epithelium. The dashed white rectangles indicate the zoomed-in regions shown in (C), bottom row.
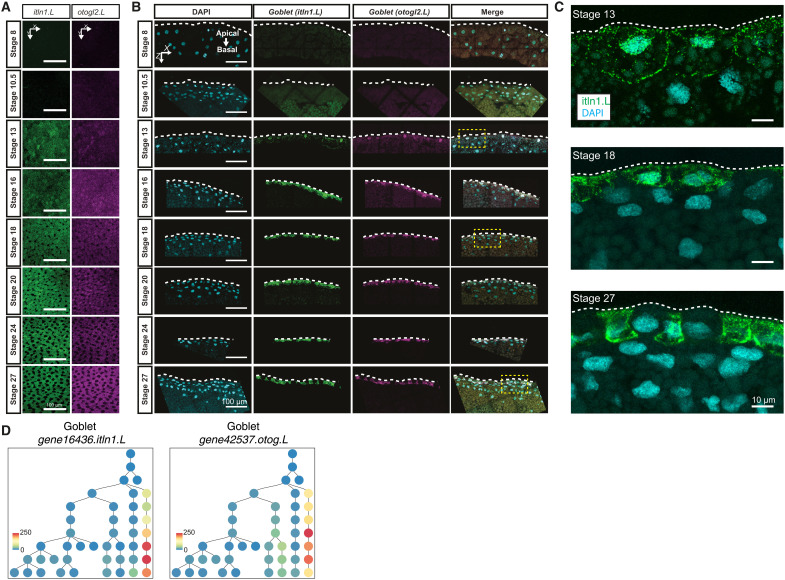
Second, the secretory transcriptional program was established exclusively through the early epithelial progenitors and their maturation, in contrast to the proposed early-stage basal cells in human and mouse cells (9, 42). The goblet-primed early epithelial progenitor lineage could be observed as early as stage 13, with a sporadic expression of marker genes (otog/mucXS/otogl2 and itln1; Fig. 5, A to C), but underwent maturation only at tailbud stages (yellow branch, light green pie; Fig. 5, A to C). Distinct from goblet cells, the ionocytes differentiated from a separate early epithelial progenitor pool by stage 16 and also underwent maturation at tailbud stages (purple branch, mixed green pie; Fig. 5, A to C). Using in situ HCR, we validated the consistent expression of ionocyte markers at stage 16 embryos (atp6v1b1: cytoplasmic; Fig. 6, A and B, and fig. S10A). Notably, in a few embryos (four of nine), we observed a few cells (presumptive Eep) that expressed ionocyte markers within single cells already at stage 13, which we confirmed both by scRNA-seq and in situ HCR (atp6v1b1; Fig. 6, A and B, and fig. S10A). The presence of these cells at stage 13 further indicates that neurula stages provide the temporal developmental window for single cells to stochastically drive multilineage programs, which, however, are executed at tailbud stages. Similarly to tekt2 in multiciliated cell progenitors, atp6v1b1 cytoplasmic expression increased within the superficial epithelium as development progressed, efficiently labeling radially intercalating ionocytes distributed in salt-and-pepper fashion next to the multiciliated cells (Fig. 6C). The developmental dynamics and TFs that drive goblet cell differentiation remain largely unresolved (Fig. 7A). Using in situ HCR, we validated the faint, interspersed expression of goblet markers in stage 13 embryos increasing over the development (otog/mucXS/otogl2 and itln1; Fig. 7, B to D, and fig. S11A, whole-mount embryos), marking both the nucleus and the cell outline. The nonuniform distribution of otogl2 and itln1 was particularly prominent within the apical surface of the superficial epithelium, outside the neuroepithelial plate (not marked by otogl2 nor itln1), at stage 13 transitioning to more homogeneous expression from stage 20 onward (Fig. 7C and fig. S11), which parallels the reported maturation of goblet cells upon regeneration (43).
Fig. 7. In situ HCR validation of developing goblet cell lineage over MCE development.
(A) In situ HCR of whole apical superficial epithelium indicating the maturation of goblet cells (itln1.L; green) and otogelin (otog.L/mucXS/otogl2.L; magenta). Dark cells (black hollow spaces) from stage 18 [here, (C)] represent radially intercalating multiciliated cells and ionocytes specified in the deep epithelial layer. Images represent the maximum intensity projections of Z sections taken from the apical superficial epithelium. (B) In situ HCR of eight developmental stages indicating goblet cell differentiation marked by intelectin (itln1.L; green) and otogelin (otog.L/mucXS/otogl2.L; magenta); nuclei are marked by DAPI staining (cyan). The yellow dashed rectangles indicate zoomed-in regions shown in (C). Images [here and in (C)] represent the maximum intensity projections of transverse cross sections, and white dashed lines indicate the apical superficial epithelium. (C) Zoomed-in view of in situ HCR marked by goblet cells (itln1.L; green) and otogelin (otog.L/mucXS/otogl2.L; magenta). At stage 13, the goblet markers are detected at very low levels with interspersed staining, but gradually mark both nuclear and cytoplasm by stage 22. (D) The timing and expression of goblet marker genes (itln1.L and otog.L) over the inferred lineage tree. The color bars indicate the scaled imputed expression of respective markers.
Third, in investigating ionocyte lineage, we classified foxi1-marked ionocytes into type I (Atp6v1highCalow) and type II (Atp6v1highCahigh), distributed in a salt-and-pepper pattern across embryonic MCE (Figs. 5, D and E, and 6, A to C, and fig. S12A) (36, 44). The ionocyte homeostasis and regeneration are regulated by cftr, and its mutations cause osmotic imbalance and modified mucus properties, leading to cystic fibrosis (9, 11). While cftr expression was missing in our single-cell dataset, we reconfirmed its patched expression in foxi1+ ionocytes using bulk transcriptomics data and in situ hybridization at tailbud stages (fig. S12, B to F) (45).
Fourth, the actively cycling basal cells (tp63 and pfn1) formed multiple subclusters (Fig. 5, A to C). The mammalian basal cells act as adult stem cells, replenishing airway epithelial cells, but their function in Xenopus MCE has remained elusive (8, 46). The in situ HCR validated basal cells (pfn1.L and tp63.L) appearance by late neurula, progressively concentrating within the sensorial layers during development (Figs. 6, A and B, and 8, A and B). The abundance of cycling basal cell subpopulations strongly suggests that the turnover of exhausted specialized cells occurs through the differentiation of late-stage basal clusters (47, 48). Notably, we observe expression of foxa1 by stage 22, labeling flat cells sitting on top of the basal cell layer expressing tp63 (Fig. 8B). Foxa1 is thought to mark the serotonin secretory cells at the tadpole stage (stage 32), and we did not observe other secretory markers. During later stages, the morphology of foxa1-expressing cells becomes rounded alongside migration toward the superficial layer (Fig. 8B), consistent with their radial intercalation into the Xenopus epidermis (25, 49).
Last, through multiplexed single-cell RNA imaging in whole embryos, we reconfirmed that the transition into gastrula and neurula stages (stages 12.5 to 13.5) is accompanied by loss of undifferentiated state marker expression (sox3.S) and the presence of early epithelial progenitors (has1.S, spdef.S, and grhl3.L) and multiciliated progenitors (mcidas.L and foxj1.L) (Fig. 8, C and D). The radially intercalating multiciliated (foxj1.L and tekt2.S) and goblet cells (grhl3.L and itln1.L) underwent respective maturation, while the basal cells within sensorial layers were marked by heterogeneous marker gene expression (tp63.L and pfn1.L) (Fig. 8, C and D).
Together, the lineage inference, in situ HCR, and multiplex RNA imaging at single-cell level provides a detailed understanding of how cell fate decisions (i.e., early epithelial progenitors) are undertaken within a temporal developmental window (i.e., neurula stages). We provide evidence that cell fate decisions, including the emergence of cell states and their underlying cellular plasticity during MCE development, are driven by a combination of molecular features (expression variance, entropy, and noise) (50, 51) and deterministic qualitative changes (changes in cell shape and migration accompanying radial intercalation) (52, 53).
Identification of early epithelial progenitors across Xenopus species
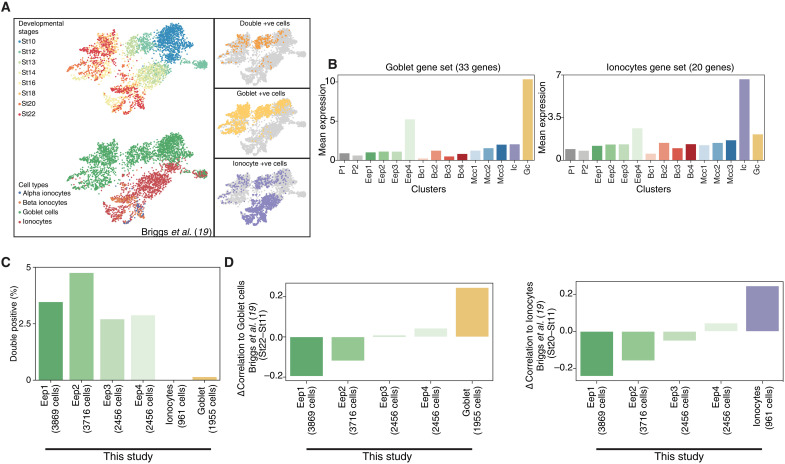
To validate that early epithelial progenitors are a bona fide cell type that drive secretory (and basal cell) lineage programs, we mined the single-cell Xenopus tropicalis developmental atlas (19), comparing the matched non-neural ectoderm lineage (stages 8 to 22) with our Xenopus laevis MCE developmental atlas (stages 8 to 27) (Fig. 9, A to D, and fig. S13, A to F). While the late-stage cell types were shared between the atlases (i.e., stage 22), the lack of early epithelial progenitors at neurula stages was puzzling. We initially correlated known cell type markers over MCE development and observed that early epithelial progenitor subclusters expressed secretory and basal markers at lower levels than late-stage cell types (fig. S14). Notably, the early epithelial progenitor subcluster (Eep4) expressed both goblet and ionocyte markers. Therefore, we classified a conservative secretory signature (33 goblet and 20 ionocytes markers) that robustly marked both cell types across both atlases (fig. S13, A and D). Using the signature gene sets, we confirmed that both mean expression and number of cells of goblet cells and ionocytes, respectively, increased over development across both studies (fig. S13, B and E). Unexpectedly, many cells within the X. tropicalis atlas expressed both secretory signatures (fig. S13, C and F), as observed with early epithelial progenitor subclusters (fig. S14). We confirmed that these double-positive cells (Ic+ and Gc+) were enriched within early neurula stages and visualized single cells in a low-dimensional expression space (Fig. 9A). Notably, the double-positive cells (Ic+ and Gc+) expressed higher levels of goblet markers (than ionocytes) and were likely classified as goblet cells. We further confirmed that early epithelial progenitors (this study) are correlated with the double-positive cells (Ic+ and Gc+) at neurula stages (X. tropicalis atlas) (Fig. 9, C and D), while the late-stage secretory cells (this study) correlate with late-stage ionocyte and goblet cells (X. tropicalis atlas) (Fig. 9, C and D). We performed in situ HCR with ionocyte (atp6v1b1) and goblet marker (otogl2) in neurula-stage embryos and found double-positive cells in both sensorial and superficial layers. The sensorial cells expressed higher levels of atp6v1b1, while superficial cells had higher otogl2, indicating distinct subpopulations (fig. S13G). In summary, MCE developmental progression requires transition through the bona fide multipotent early epithelial progenitors before differentiation into the secretory cell types, and the early epithelial progenitors have likely been misannotated owing to the variable expression of multiple secretory markers.
Fig. 9. Function of multipotent early epithelial progenitors in the development of the secretory cell types.
(A) Low-dimensional visualization of non-neural ectoderm cells, stages, and their annotations across X. tropicalis development atlas. The insets indicate the goblet cells, ionocytes, and double-positive cells marked by core signature gene sets. (B) Mean expression of goblet (33 genes) and ionocyte (20 genes) signature genes over MCE developmental clusters (this study). While late-stage goblet cells and ionocyte have the highest mean expression of signature gene sets, the early epithelial progenitor clusters also express both signature genes. (C) Percentage of early epithelial progenitors expressing both goblet (33 genes) and ionocyte (20 genes) signature genes. Nearly all double-positive cells are early epithelial progenitors. (D) Change in the correlation between stage 22 and stage 11 goblet cells and ionocytes across X. tropicalis development atlas and this study. The correlation at stage 11 (early stages) is higher than early epithelial progenitors, while at late stage (stage 22), correlation indicates respective goblet cells and ionocytes.
Evolutionary conservation of MCE cell types
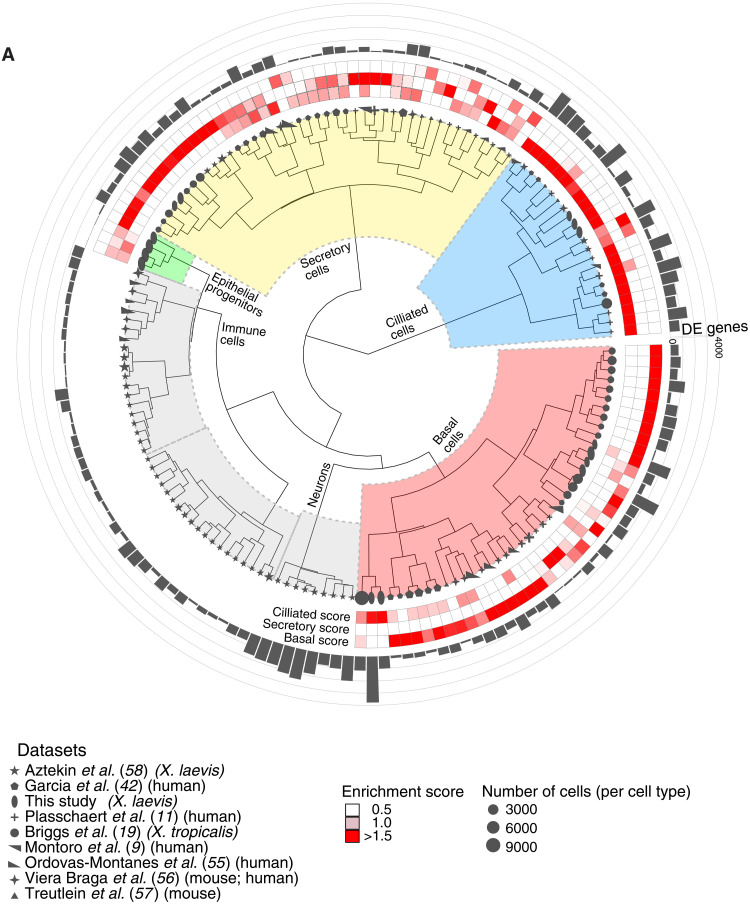
Recently, many cell types of the Xenopus MCE and their functional equivalents have been identified during airway development in mouse and humans, aided by single-cell transcriptomics (9–12, 54). We investigated whether the transcriptional programs (i.e., expression modules) were conserved across vertebrates and drive cell type function. To characterize the conservation and divergence of MCE cell types, we performed a comparative analysis of our data with eight single-cell transcriptomics atlases spanning X. laevis (adult epidermal regeneration), X. tropicalis (embryonic and adult epithelium), mouse trachea, and human nasal epithelium (9, 11, 19, 42, 55–58), spanning 144 cell types across 120,842 single cells (table S12). To compare cell types across different species, we reanalyzed all single-cell atlases in a standardized and uniform manner, retaining respective author annotations (gray shapes; Fig. 10A). We devised a differential expression-based cell type enrichment score (Fig. 10A and Materials and Methods) for each cell type [accounting for the number of cells; black rectangular bars indicate differentially expressed (DE) genes; Fig. 10A and Materials and Methods] that robustly distinguished the secretory, ciliated, and basal transcriptional cell types from other cell types, using author-specified cell types as ground truth (Fig. 10A; fig. S15, A to D; note S4; and Materials and Methods). The “ciliated,” “secretory,” and “basal” enrichment scores (red squares; Fig. 10A) were calculated for each cell type (across nine single-cell atlases), indicating the specificity of marker genes to classify respective cell types (fig. S15A), enabling a comparative analysis between species. We observed the highest DE genes (and respective scores) in ciliated cell types, followed by basal and secretory cell types (enrichment scores in red squares; the number of DE genes are in gray bars; Fig. 10A). The secretory cell types (goblet cells, ionocytes, and subtypes) from three Xenopus atlases were grouped together but separated from their mouse and human counterparts [Tuft, Brush, Pulmonary neuroendocrine cells (PNEC), etc. cell types] (gray shapes; Fig. 10A and fig. S15B). The Xenopus goblet cells lack canonical mucins present in higher vertebrates, while ionocytes were originally characterized in Xenopus (36) and their disease-related counterparts have been identified in the mammalian airway (9, 11). Similar to the secretory program, the mouse and human specialized basal cell types were grouped separately from both Xenopus counterparts (Fig. 10A and fig. S15C), indicating a distinct higher vertebrate basal expression program. Notably across ciliated cells, we observed a conserved transcriptional program of ciliated progenitors across species (two mouse ciliated progenitors and three Xenopus ciliated progenitors) and a clear distinction of mature ciliated cell types (multiple mouse, human, and Xenopus cell types; Fig. 10A and fig. S15D), indicating that paradigms of ciliogenesis and maturation are conserved across higher vertebrates. We highlight enriched genes and their expression in different cell types across species (fig. S15E). In summary, the comparative analysis captured the functional importance and conserved transcriptional programs during ciliogenesis across vertebrates, while the regulation of cellular identity across secretory and basal cell programs is driven by cell type–specific programs.
Fig. 10. Comparative analysis of MCE cell types and development.
(A) Comparative analysis between Xenopus, mouse, and human mucociliary cells from nine single-cell airway atlases, using expressed orthologous gene sets. Using author-annotated cell type labels (shapes), a cell type enrichment score (ciliated, secretory, and basal; enriched scores in red) is calculated on the basis of differentially enriched genes (gray bars), which classified the different cell types into basal, ciliated, secretory, and other cell types (immune and neuronal). The four early epithelial progenitor clusters together with secretory cell types. The shapes indicate the respective study, shape sizes indicate the scaled number of cells within each author-annotated cell type, and gray histograms indicate the number of differentially enriched genes per author-annotated cell type. The legend further indicates the respective study, organism type, and species; coloring of enrichment score and sizes indicates the number of cells per author-annotated cell type.
DISCUSSION
Investigating the composition, development ordering, and dynamics of MCE cell types during embryogenesis is of fundamental importance to vertebrate biology. The Xenopus mucociliary epidermis enables an understanding of mucociliary development without the prerequisite of lung formation and provides a critical developmental and evolutionary perspective to comprehensive studies in mammals focusing on MCE regeneration and homeostasis.
Here, we profiled functional MCE formation across 10 developmental stages at a single-cell resolution and captured transitory and mature cell types over the MCE development that lack discrete bifurcation points. Our data highlight that homogeneous blastula undergoes the key initial developmental transition to the multiciliated progenitors and a multipotent early epithelial progenitor ensemble that drives the body plan (gastrula and neurula) and subsequent organogenesis in trajectories to specialized cell types during tailbud stages. We report that Xenopus MCE differentiation follows a continuous nonhierarchical developmental model, which is neither discrete nor completely stochastic. We speculate that the two gastrula subpopulations represent the two-layered epidermal cells with varied plasticity in organoids and embryos. Furthermore, in this study, we uncovered the initial fate transition, alongside expression changes in TFs and marker genes, involved in the complex coordination of signaling, cell cycle, and spatiotemporal cell type organization. The expression dynamics over the branched developmental manifold and pseudotime highlight that diverging multilineage programs occur at different developmental speeds. This dynamic equilibrium of progenitor specialization is critical for maintaining signaling and maternal gradients, as the depletion of either multiciliated, ionocytes, basal, or goblet cells leads to failures in the formation of terminal cell type and mucociliary functions (24, 59). Through radial intercalation, multiciliated precursors migrate from the sensorial layer to embed between secretory cells at superficial epithelial layers (41). Alongside the maturation of multiciliated cells, we capture and validate early epithelial progenitors as the precursors for basal and secretory cellular programs. We observed heterogeneity in basal cell clusters, based on not only marker genes but also cell cycle proportions, which can contribute to regenerating adult MCE cell types, as in mammals (8, 46). We uncover basal cell subpopulations, which retain the developmental plasticity to further differentiate and potentially replenish adult MCE cell types. The Xenopus mature cell types have recently been reported to undergo transdifferentiation to other cell types (60). We speculate that this fate conversion could be facilitated through the early epithelial progenitors and subsequent basal cell subpopulations that retain cellular plasticity and maintain MCE composition in homeostasis and regeneration (60, 61).
Through MCE development, we observe stochastic gene expression within neurula-stage progenitors contributing to the expression of multilineage expression programs, resolving into late-stage cell type–specific gene regulatory networks. The molecular features (decreasing transcriptional diversity, entropy, and increased molecular noise) coupled with cell shape and migration indicate that neurula stages provide the developmental window to establish initial heterogeneity and cell fate switching frequencies, which are resolved by late-stage maturation into specific cell types (39, 40). Our single-cell in silico lineage inference coupled with in situ HCR validation and single-cell multiplexed RNA imaging captured the timing, speed, and maturation of secretory cell types and their transcriptional program across early epithelial progenitor lineages, while multiciliated progenitors originated from pluripotent cells and underwent maturation in a separate branch. Notably, we find early epithelial progenitors also present across X. tropicalis non-neural ectoderm cells, expressing both goblet cell and ionocyte signatures. The X. tropicalis developmental atlas spans all germ layers, sampling many more cells, while this study is focused on the sampling of developing MCE and its cell types. Within the double-positive secretory cell types at neurula stages, the goblet markers were expressed at higher mean levels (relative to ionocyte markers) and were likely to have confounded the annotation and absence of early epithelial progenitors. The presented lineage inference uses expression data alone, without additional priors and assumes unidirectional progression without trans/dedifferentiation. Future versions could incorporate spatial positioning, cell shape dynamics, and cell cycle, including growth and division rates. Our data and lineage inference provide a quantitative and generalizable approach to characterizing a continuous model of cell fate choice suited for temporal data, uncovering general features of embryonic MCE development.
We also find the type I and type II ionocytes that are functional equivalents of human alpha and beta submucosal epithelial serous acinar cells controlling ion flow, regulating fluid and bicarbonate secretion to hydrate mucins, and balancing acid-base homeostasis and thereby the volume and composition of the air-surface liquid. Although human primary models (air-liquid interface, human airway epithelial culture) are valuable, they are technically demanding and do not fully recapitulate relevant cell type interactions occurring in vivo. Several MCE cell types and hallmarks are also found in other tissues, including the pronephric kidney, gut, epididymis, and nasal cavity across species, with conserved TF families and signaling pathways. It is tempting to speculate that similar paradigms and gene regulatory networks exist across other MCE tissues to drive cell fate decisions. Through comparative analysis across nine single-cell datasets, we identified conserved transcriptional programs driving ciliogenesis in cell types across vertebrates, while specialized cell types and their cellular functions drive specific transcriptional programs across secretory and basal cell types. We infer the regulatory relationships during MCE development through scRNA-seq; however, future studies that apply single-cell multiomic approaches (i.e., chromatin accessibility, transcriptomics, spatial positioning, etc.) will help integrate signaling regulation, transcription factor enhancer cross-talk, further to push an improved understanding of cell fate decisions.
The current study provides insights into MCE development; however, there are some important considerations of note. First, we leverage Xenopus animal cap organoids that mimic embryonic MCE development but have distinct commonalities and differences with the embryo. Second, while the highly abundant transcripts are robustly captured in the scRNA-seq data, many of the lowly expressed TFs signaling components are not missed in the current analysis and further lost during ortholog mapping to human identifiers. Third, we observe expression of foxa1, a canonical marker of serotonin secretory cells, within single cells at stage 22. These cells lack the expression of other serotonin secretory markers, which is likely because of limited sequencing depth, but require further characterization to appreciate the contribution of the later developmental stages. Last, the insights from scRNA-seq analysis in this study are supported by in situ HCR and multiplex spatial RNA imaging. We hope that further functional studies will help understand the mechanistic insights into the development and species-specific differences across mucociliary epithelia.
Our work provides a resource of cell state changes and developmental transitions accompanying MCE development and contributes to dissecting developmental mechanisms involved in the formation of mucociliary tissues. The identified and conserved molecular targets, mechanisms, and cell fate paradigms controlling MCE differentiation would be useful in the development of regenerative therapies, including ciliopathies and chronic lung diseases, aimed at restoring mucociliary clearance.
MATERIALS AND METHODS
Experimental methods
All reagents and resources referred to below are summarized with vendors and identifiers in table S11.
X. laevis husbandry, embryo isolation, and manipulation
Wild-type X. laevis were obtained from Nasco, Wisconsin, Fort Atkinson, WI, USA. Frogs were maintained by centralized facilities according to the common procedures provided by the international Xenopus community including National Xenopus Resource at the Marine Biological Laboratory, Woods Hole, USA; the European Xenopus Resource Center at the University of Portsmouth, School of Biological Sciences, UK; and also Xenbase (http://xenbase.org). X. laevis embryos were isolated and manipulated according to standard procedures: X. laevis females were ovulated by injection of 500 U of human chorionic gonadotropin. The following day, eggs were collected, in vitro fertilized, and dejellied in 2% cysteine (pH 7.9). Embryos were washed and reared in 1/3× Marc’s modified Ringer’s solution. Embryos were incubated until stage 8 according to NF developmental tables (23).
Animal cap culture, single-cell dissociation, and transcriptomics
A total of 100 X. laevis animal cap from at least three independent embryo clutches were manually dissected from embryos at NF stage 8 as described in (62) and were subsequently cultured in Danilchik’s for Amy medium supplemented with gentamycin (50 μg/ml) until the desired developmental stage was assessed by comparing to sibling embryos. From each of the 10 sampled embryonic stages (NF stages 8, 10.5, 12, 13, 16, 18, 20, 22, 24, and 27), animal cap organoids were dissociated into a single-cell suspension by first washing with 0.01% bovine serum albumin–phosphate-buffered saline (BSA-PBS) and then incubating with the Newport buffer 2.0 described in (19) in a BSA (1 mg/ml)–pretreated 1.5-ml Eppendorf at room temperature (RT) while agitating on a thermomixer, typically about 5 min at 500 rpm for animal cap NF stages 8 to 22 and additional 10 min at 1200 rpm for NF stages 24 and 27. The single-cell suspension was spun down and washed with 0.01% BSA-PBS twice at 300 relative centrifugal force (rcf) for 3 min and then spun down at 300 rcf for 5 min and resuspended in 0.01% BSA-PBS. Cells were passed through a 50-μm-diameter cell strainer, and the concentration was adjusted to 500 cells/μl. We evaluated the quality and purity of single cell in suspension by microscopy by staining with propidium iodide (2 μg/ml) and 20-μm Hoechst 33342 and found negligible cell death (<1%) up to 1 hour after animal cap organoid dissociation. The single-cell suspension was left on ice for 15 min, before processing and loading using a wide bore tip on the 10x Chromium chip for droplet-based scRNA-seq v2 (stages 8, 13, 16, 18, 20, 24, and 27) and v3 (stages 10.5, 12, and 22), following the manufacturers’ recommended protocol (63), and sequenced using paired-end reads on an Illumina NextSeq 500 (high-output reagent kit v2.5, 75 cycles) for 90 cycles with read 1 [26 base pairs (bp), 10× bead barcode and unique molecular identifiers (UMIs)], read 2 (56 bp, transcript), and index read (8 bp, 10× sample index).
Whole-mount in situ hybridization and HCR
The X. laevis embryos were in situ hybridized with Cftr.L probes according to Lukoseviciute et al. (64). For in situ HCR on whole X. laevis, embryos were prepared as for a traditional in situ hybridization, up to the probe hybridization step, as described in (64) followed by the in situ HCR protocol described in (61). Hybridization probes were designed and generated by Molecular Instruments Inc. Embryos were fixed with 4% formaldehyde in 1× MOPS, EGTA, MgSO4, and Paraform Fixative (MEMFA) salts in Wheaton vials for 60 min on a rotator at RT, washed 3× for 15 min with PBS, and permeabilized with 100% methanol overnight at −20°C. The following day, embryos were rehydrated by 5 min of wash in (i) 75% methanol/25% H20, (ii) 50% methanol/50% H20, and (iii) 25% methanol/75% H20 and then washed two times for 5 min in 100% Phosphate-buffered saline/Tween (PBST). Next, embryos were permeabilized with proteinase K (final concentration of 5 μg/ml in PBST) for 10 min at RT. Embryos were washed three times for 5 min in PBST to remove proteinase K and washed two times for 5 min in 0.1 M triethanolamine. After that, 10 μl of acetic acid anhydride was added to 0.1 M triethanolamine, and embryos were incubated for 10 min at RT. Embryos were washed two times for 5 min with PBST and refixed in 4% formaldehyde in H20 for 20 min at RT on a rotator. Next, embryos were washed five times for 5 min with PBST. Embryos were washed for 5 min in a preheated (37°C) 500-μl wash buffer (Molecular Instruments Inc.) and transferred to 1.5-ml Eppendorf tubes (10 embryos per tube). The washed buffer was removed and replaced by a preheated 500-μl hybridization buffer (Molecular Instruments Inc.) for a 30-min incubation at 37°C. The hybridization buffer was removed and replaced by a 500-μl probe solution prepared by diluting a probe stock in the hybridization buffer to the final concentration of 6 nM. Subsequently, embryos were incubated at 37°C overnight, washed two times for 30 min with a wash buffer (Molecular Instruments Inc.), washed for 5 min with 50% 5× Saline-sodium citrate/Tween (SSCT)/50% wash buffer for 5 min, and two times for 20 min with 5× SSCT at RT. Embryos were preamplified with 1 ml of amplification buffer (Molecular Instruments Inc.) for 10 min at RT. To visualize the probes, the preamplification buffer was replaced by an amplification solution prepared by first heating to 95°C the fluorescently tagged hairpins pairs (h1 and h2 hairpins) corresponding to the designed probes. Hairpins were incubated for 30 min at RT in the dark and subsequently added to 500 μl of amplification buffer (Molecular Instruments Inc.) and incubated for 5 min in the dark. The final concentration of hairpins was 48 nM. The preamplification buffer was removed from embryos and replaced by the hairpin solution. Embryos were incubated overnight at RT in the dark. The following day, embryos were transferred to Wheaton vials and washed two times with 5× SSCT for 30 min and stored in 1× PBS at 4°C. For immunostaining after in situ, embryos were blocked in 1 ml of 50% CAS-Block/50% PBST for 1 hour in Wheaton vials at RT in the dark followed by incubation with the primary antibody (anti-acetylated tubulin; 1:500; Sigma-Aldrich, T7451) in CAS-Block at 4°C overnight in the dark. The next day, embryos were washed three times for 10 min with PBST and incubated with a secondary antibody (anti-mouse 647; 1:250; Thermo Fisher Scientific, A21235) diluted in 1 ml of 50% CAS-Block/50% PBST for 1 hour at RT. Embryos were washed three times for 10 min with PBST in the dark, mounted, and imaged using a Zeiss 880 inverted confocal microscope using C-Apochromat 40×/1.2 W Corr M27 objective. For sectioning, embryos were embedded in 2% low–melting point agarose, sectioned using a vibratome set up to a 50-μm section thickness, stained with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml; Thermo Fisher Scientific, D1306) in 1× PBS at RT for 10 min, mounted in Moviol [10% Mowiol 4-88, 25% glycerol, 0.1 M tris (pH 8.5), and 2% N-propyl gallate] with coverslip, and imaged using a Zeiss 880 inverted confocal microscope using C-Apochromat 40×/1.2 W Corr M27 objective. Fiji (65) was used for image processing including the maximum intensity projection of the acquired z-stacks and contrast adjustment.
Molecular cartography
Sample fixation
X. laevis embryos were fixed and stabilized according to Resolve guidelines. Briefly, embryos were incubated at RT in PAXgene fixation solution for 4 hours followed by 48 hours of incubation at RT in a PAXgene stabilizer. Next, embryos were incubated for 48 hours in 20% sucrose (w/v in 1× PBS) at 4°C until embryos sank to the bottom of the well.
Cryo-embedding and sectioning
Embryos were washed of 20% sucrose (w/v in 1× PBS) in 1× PBS/OCT (optimal cutting temperature medium; Neg-50 Frozen Section Medium, VWR) (2:1, 1:1, 1:2, and 100% OCT) for 30 min each wash at RT. Next, embryos were placed in the middle of the cryo-embedding medium. The cryomold was placed on top of the liquid nitrogen in the gas phase until the medium was completely white. The cryo-embedded block was kept in dry ice until sectioning. Embryos were sectioned at 12 μm, and sections were collected onto a precooled coverslip. The samples were sent to Resolve GmbH, Germany for processing.
Molecular cartography
PAXgene fixed samples were used for Molecular Cartography according to the manufacturer’s instructions (protocol 3.0; available for download from Resolve’s website to registered users), starting with the aspiration of ethanol and the addition of buffer BST1 (steps 6 and 7 of the tissue priming protocol). Briefly, tissues were primed followed by overnight hybridization of all the probes specific for the target genes (see below for probe design details and target list). Samples were washed the next day to remove the excess probes and were fluorescently tagged in a two-step color development process. Regions of interest were imaged as described below, and fluorescent signals were removed during decolorization. Color development, imaging, and decolorization were repeated for multiple cycles to build a unique combinatorial code for every target gene that was derived from raw images as described below.
Probe design
The probes for selected genes were designed using Resolve’s proprietary design algorithm. Briefly, the probe design was performed at the gene level. For every targeted gene, all full-length protein-coding transcript sequences from the ENSEMBL database were used as design targets if the isoform had the GENCODE annotation tag “basic” (66). To speed up the process, the calculation of computationally expensive parts, especially the off-target searches, the selection of probe sequences was not performed randomly but was limited to sequences with high success rates. To filter highly repetitive regions, the abundance of k-mers was obtained from the background transcriptome using Jellyfish (67). Every target sequence was scanned once for all k-mers, and those regions with rare k-mers were preferred as seeds for a full-probe design. A probe candidate was generated by extending a seed sequence until a certain target stability was reached. A set of simple rules was applied to discard sequences that were found to experimentally cause problems. After these fast screens, every kept probe candidate was mapped to the background transcriptome using ThermonucleotideBLAST (67), and probes with stable off-target hits were discarded. Specific probes were then scored on the basis of the number of on-target matches (isoforms), which were weighted by their associated annotating principal splice isoforms (APPRIS) level (68), favoring principal isoforms over others. A bonus was added if the binding site was inside the protein-coding region. From the pool of accepted probes, the final set was composed by greedily picking the highest-scoring probes.
Imaging
Samples were imaged on a Zeiss Celldiscoverer 7, using the 50× Plan Apochromat water immersion objective with a numerical aperture of 1.2 and the ×0.5 magnification changer, resulting in a ×25 final magnification. Standard CD7 light-emitting diode excitation light sources, filters, and dichroic mirrors were used together with customized emission filters optimized for detecting specific signals. The excitation time per image was 1000 ms for each channel (DAPI was 20 ms). A z-stack was taken at each region with a distance per z-slice according to the Nyquist-Shannon sampling theorem. The custom CD7 complementary metal-oxide semiconductor camera (Zeiss Axiocam Mono 712; 3.45-μm pixel size) was used. For each region, a z-stack per fluorescent color (two colors) was imaged per imaging round. A total of eight imaging rounds were done for each position, resulting in 16 z-stacks per region. The completely automated imaging process per round (including water immersion generation and precise relocation of regions to image in all three dimensions) was realized by a custom python script using the scripting API of the Zeiss ZEN software (open application development).
Spot segmentation
The algorithms for spot segmentation were written in Java and are based on the ImageJ library functionalities. Only the iterative closest point algorithm is written in C++ based on the libpointmatcher library (https://github.com/ethz-asl/libpointmatcher).
Preprocessing
As a first step, all images were corrected for background fluorescence. A target value for the allowed number of maxima was determined on the basis of the area of the slice in square micrometer multiplied by the factor 0.5. This factor was empirically optimized. The brightest maxima per plane were determined on the basis of an empirically optimized threshold. The number and location of the respective maxima was stored. This procedure was done for every image slice independently. The maxima that did not have a neighboring maximum in an adjacent slice (called z-group) were excluded. The resulting maxima list was further filtered in an iterative loop by adjusting the allowed thresholds for (Babs-Bback) and (Bperi-Bback) to reach a feature target value (Babs: absolute brightness; Bback: local background; Bperi: background of periphery within 1 pixel). This feature target values were based on the volume of the three-dimensional (3D) image. Only maxima still in a z-group of at least two after filtering were passing the filter step. Each z-group was counted as one hit. The members of the z-groups with the highest absolute brightness were used as features and were written to a file. They resemble a 3D point cloud.
Segmentation and decoding
To align the raw data images from different imaging rounds, images had to be corrected. To do so, the extracted feature point clouds were used to find the transformation matrices. For this purpose, an iterative closest point cloud algorithm was used to minimize the error between two point clouds. The point clouds of each round were aligned to the point cloud of round one (reference point cloud). The corresponding point clouds were stored for downstream processes. On the basis of the transformation matrices, the corresponding images were processed by a rigid transformation using trilinear interpolation.
The aligned images were used to create a profile for each pixel consisting of 16 values (16 images from two color channels in eight imaging rounds). The pixel profiles were filtered for a variance from zero normalized by the total brightness of all pixels in the profile. Matched pixel profiles with the highest score were assigned as an ID to the pixel. Pixels with neighbors having the same ID were grouped. The pixel groups were filtered by group size, number of direct adjacent pixels in the group, and number of dimensions with a size of two pixels. The local 3D maxima of the groups were determined as potential final transcript locations. Maxima were filtered by the number of maxima in the raw data images where a maximum was expected. The remaining maxima were further evaluated by the fit to the corresponding code. The remaining maxima were written to the results file and considered to resemble transcripts of the corresponding gene. The ratio of signals matching to codes used in the experiment and signals matching to codes not used in the experiment were used as estimation for specificity (false positives).
Computational analysis
Single-cell data alignment and quality control
The raw 10x Genomics sequencing data were processed using the Cell Ranger (version 2.1.1) with the default option for 3′ Gene Expression v2 Libraries (“SC3Pv2”). The sequences were mapped to X. laevis genome (XenBase RRID# SCR_003280; NCBI GCA_001663975.1) using the annotation from the GitLab repository (https://gitlab.com/Xenbase/genomics/XENLA_9.2; 2018-May version) (45, 69). The 10 individual scRNA-seq datasets were aligned to the reference genome (XENLA_GCA001663975v1_XBv9p2) using zUMI’s alignment and feature counting (version 2.4.5b), which uses Spliced Transcripts Alignment to a Reference (STAR) version 2.5.4b (70, 71). The XENLA 9.2 assembly included two annotations for gene otogelin (mucXS) referred as otog and otogl2. To filter out background barcodes, we applied EmptyDrops (from package DropletUtils version 1.2.2) using default parameters and selected only cells passing a < 0.01 false discovery rate threshold for every individual dataset (72). To identify doublets, we applied scrublet (version 0.2.1) using parameters expected_doublet_rate = 0.06, min counts = 2, min_cells = 3, min_gene_variability_pctl = 85, and n_prin_comps = 30 to all datasets individually (72, 73). We normalized the datasets for cell library size, log-transformed and scaled genes to mean zero and unit variance, and identified HVGs using scanpy (version 1.4) (74).
Data visualization and marker gene detection
For each MCE developmental stage, we performed dimensionality reduction using uniform manifold approximation and projection (UMAP) (75) with 10 neighbors on 40 principal components and identified clusters using Louvain community detection. To generate an integrated force-directed k-nearest neighbor (knn) graph layout, we applied SPRING to the integrated single-cell expression data using default parameters (76). We performed a Wilcoxon rank sum test between each identified cluster and the remaining cells in the integrated dataset using the scanpy function scanpy.tl.rank_genes_groups with use_raw = True.
Imputation
We imputed gene counts using Markov affinity-based graph imputation of cells (MAGIC) (using palantir.utils.run_magic_imputation) using parameters n_steps = 3 (76). Note that imputed values were only used for visualization of marker gene expression of embeddings and to infer gene trends.
Developmental dynamics
To infer pseudotime and developmental potential, we applied Palantir (version 0.2.1) using default parameters (77). To set the start cell, we selected one arbitrary cell from the earliest sample time point. To run Palantir on discrete time series data, we “stitched” it together with harmony (version 0.1.1) (78) using harmony.core.augmented_affinity_matrix with parameters n_neighbors = 20 and pc_components = 50. Diffusion maps (also from the Palantir package) were computed with parameters n_components = 20 and knn = 20. To validate the inferred developmental dynamics, we applied CytoTRACE (version 0.1.0) using the function iCytoTRACE for dataset integration with default parameters (40).
PhenoGraph
To cluster the integrated dataset, we used PhenoGraph (version 1.5.2) (76) through the Palantir function Palantir.utils.determine_cell_clusters with parameter k = 1000.
Louvain
Individual stages were clustered using louvain, through scanpy.tl.louvain, using 10 neighbors and 40 principal components. Louvain resolution parameters were set for each stage individually (in range 0.01 to 0.3).
Cluster stability
Robustness of clustering was assessed by repeating 20 independent iterations of PhenoGraph and Louvain clustering for each developmental stage 20 times, using different random seeds and calculating element-wise clustering consistency (element_consistency) using the ClustAssess package (version 0.3.1) with default parameters (79).
Gene trends
Gene trends over branches were computed using the Palantir function palantir.presult.compute_gene_trends on the MAGIC-imputed gene counts (76).
Gene ontology
We use ClusterProfiler (version 3.10.1) for GO analysis (80). X. laevis gene names were mapped to human orthologs. We used enrichGO per stage to cluster gene sets to assert the statistically significant enriched biological processes. Comparison between individual stages or clusters were achieved using the compareCluster function. Statistically significant terms were selected with a P < 0.05 threshold, after Benjamini-Hochberg correction for multiple testing.
Lineage inference
To infer the developmental lineages, we applied a neighbor mapping and voting strategy. Here, each cell votes for the cluster of its nearest neighbor at the preceding time point. Votes are aggregated within clusters and a link is drawn to the most likely developmental “ancestor.” Neighbor search is performed with the Scipy (version 1.1.0) function scipy.spatial.cKDTree using default parameters. Upon assessment of voting confidence and marker gene expression, low-confidence links were refined using the expression of key branch marker genes.
Gene set quantification
To quantify the utilization of different functional gene groups, we plotted the normalized and z-scored gene expression of the cell cycle, TF, and signaling gene sets, retrieved from public databases.
Cell cycle
Cell cycle genes were retrieved from Cyclebase (81).
Transcription factors
Human TFs were retrieved from AnimalTFDB 3.0 (37).
Signaling
Signaling gene sets were obtained from MSigDB (gsea-msigdb.org/) and converted to X. laevis orthologs. A handful of X. laevis gene sets were removed (hvl and hdac from the Notch pathway; smad2/3/4 from the Wnt pathway; and rho, rock1/2, and e2f4/5 from the TGF-β pathway) and manually added (wnt, fzd, and lrp5 to the Wnt pathway and tgfbr1, gdf11.1, and nodal inhbc2 to the TGF-β pathway).
Cell cycle classification
The cell cycle stage prediction was performed using the scanpy function (scanpy.tl.score_genes_cell_cycle) to score S and G2-M phase genes. The reference cell cycle stage marker genes can be found at https://github.com/theislab/scanpy_usage/blob/master/180209_cell_cycle/data/regev_lab_cell_cycle_genes.txt. Each single cell is assigned an S and G2-M score and is classified on the basis of the highest-scoring gene set. If neither gene set scores above 0.5, then the cell is classified as G1.
RNA velocity
On the zUMI’s generated intron-exon classified output, we applied scVelo (version 0.2.2) (33) per stage and branch. scVelo was run on 10 principal components and 50 neighbors, in “stochastic” mode.
Bulk Xenopus dataset mapping
Expression data were retrieved through GEO (GSE76342). Gene names were mapped to X. laevis gene names. If several genes map to a X. laevis gene name, then only the first is retained. To assess similarity, we performed Spearman correlation between the two datasets, on all HVGs also detected in (38), totaling 1376 genes.
X. tropicalis cell type mapping
The non-neural ectoderm lineage cells were reanalyzed from X. tropicalis single-cell developmental atlas (19) and compared to our cell types. A gene set consisting of 33 goblet markers and 20 ionocyte markers was selected, which robustly marked the respective cell types across both studies (based on author annotation). The mean expression of the respective goblet cell and ionocyte gene sets was used to reclassify single cells as single-positive (Gc+ or Ic+) or double-positive cells (Ic+ and Gc+) from the non-neural ectoderm lineage from all stages.
Single-cell evolutionary development comparison
We downloaded eight single-cell datasets spanning Xenopus, mouse, and human nasal/airway epithelium atlases from respective repositories, spanning 144 cell types across 120,842 single cells (note S4). We converted all gene symbols to human orthologs and used them as a common reference to project datasets onto each other. For each dataset and respective cell types, marker genes were identified first using scanpy (P < 0.05). All marker genes from all cell types within each data were combined to form dataset-specific gene sets, and enrichment analysis (per cell-type) was calculated, i.e., the mean expression in a given cell type relative to the mean expression within the dataset. The individual cell type gene enrichments were pairwise-correlated (rank-based Spearman correlation) within and across datasets. To define the basal, secretory, and multiciliated gene sets, we used differential expressed genes identified in at least 20% of cell types within each cluster. Each cell type was subsequently scored (enrichment score) on the basis of gene expression similarity to the conserved gene sets within the clusters of cell types. The conserved marker genes (fig. S15E) were identified by counting the number of times a given gene was classified as a marker and ranked based on frequency, i.e., how often genes were classified as markers in either multiciliated, ionocyte, or goblet cell types and across all datasets.
Multiplexed single-cell spatial analysis
The nuclear segmentation of cells was performed using the manufacturer’s recommendations, i.e., the maximum intensity projection using watershed cell detection in QuPath (version 0.2.3) with parameters (background radius = 25 pixels, median filter radius = 6 pixels, sigma = 8 pixels, minimum area = 25 pixels2, maximum area = 6000 pixels2, threshold = 30, cell expansion = 50 pixels, and smooth boundaries enabled). Spots were quantified in each region of interest using Polylux version 1.6.1 (Resolve Biosciences) within Fiji/ImageJ2 version 2.1.0/1.53c. Individual segmented cells with less than eight spot counts were disregarded, and spot counts were normalized to the same number of spots per cell to facilitate comparison between cells. Normalized spot counts (more than eight spots) for marker gene probes were superimposed over DAPI-segmented nuclei and sectioned images. The relative marker gene expression across stages was calculated for all cells passing the quality control, followed by single-cell classification into either multiciliated, goblet, basal cells, or ionocytes. The heatmap was plotted using a scaled mean expression across all stages.
Statistics and reproducibility
Animal cap organoids were derived from at least three different X. laevis females.
Ethics declarations
This work was done in compliance with Danish animal protection laws and was approved under animal license 2017-15-0201-01237 and project plan P21-124.
Acknowledgments
We thank C. Aztekin for in situ HCR guidance, as well as H. Neil, M. Michaut (reNEW Genomic Platform), M. Mechta and the Single-Cell Omics Platform at the Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR), and J. Bulkescher (reNEW Imaging Platform) for technical expertise, support, and help. We also thank members of the Sedzinski, Kwon, and Natarajan groups. We thank DeIC National HPC Centre (ABACUS 2.0 and eScience Center) for computational resources. We also thank J. B. Wallingford for discussions and comments on the manuscript.
Funding: The research in the KNN lab is supported by a Villum Young Investigator grant (VYI#00025397) and a Novo Nordisk foundation grant (#NNF18OC0052874). The Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW) is supported by Novo Nordisk Foundation grant nos. NNF17CC0027852 and NNF21CC0073729. J.S., J.L., and A.B. acknowledge the Novo Nordisk Foundation (NNF19OC0056962) and Leo Foundation (LF-OC-19-000219) for funding support. T.K. and S.C. are supported by the UNIST Future Project Research Fund (1.220023.01) and NRF-Basic Science Research Program (2018R1A6A1A03025810). A.F.M. and K.N.N. acknowledge the Sino-Danish Center and Louis Hansen Foundation (J.nr.20-2B-6705) for funding support. Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center, based at the University of Copenhagen, and partially funded by an unconditional donation from the Novo Nordisk Foundation (grant no. NNF18CC0034900).
Author contributions: J.S., J.L., and A.B. collected tissue samples and performed scRNA-seq. T.H.P. contributed to sequencing libraries. J.S. and A.B. performed in situ HCR. J.S., A.B., T.K., Y.K., H.-S.L., and T.J.P. contributed toward experimental validation. A.F.M. and K.N.N. processed and analyzed all data. S.C. and T.K. also contributed to data analysis. K.N.N. wrote the manuscript with input from co-authors. J.S., T.K., and K.N.N. supervised the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The sequencing data and processed gene counts are available on GEO (accession number GSE158088). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw data alongside analysis scripts are available at Zenodo (10.5281/zenodo.7602447) and GitHub (https://github.com/Natarajanlab/XenopusMCEAtlas). The single-cell transcriptomics data and metadata can be downloaded for offline visualization or can be interactively visualized at CellxGene (https://cellxgene.cziscience.com/collections/d4055728-b22d-4851-b12d-d7bd0216e8c4) or UCSC Cell Browser (https://cells-test.gi.ucsc.edu/?ds=xenopus-dev).
Supplementary Materials
This PDF file includes:
Notes S1 to S4
Figs. S1 to S15
Legends for tables S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S12
REFERENCES AND NOTES
- 1.P. Walentek, Manipulating and analyzing cell type composition of the xenopus mucociliary epidermis. Methods Mol. Biol. 1865, 251–263 (2018). [DOI] [PubMed] [Google Scholar]
- 2.J. M. Hayes, S. K. Kim, P. B. Abitua, T. J. Park, E. R. Herrington, A. Kitayama, M. W. Grow, N. Ueno, J. B. Wallingford, Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev. Biol. 312, 115–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. R. Rock, X. Gao, Y. Xue, S. H. Randell, Y.-Y. Kong, B. L. M. Hogan, Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639–648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. C. Nawroth, A. M. van der Does, A. Ryan Firth, E. Kanso, Multiscale mechanics of mucociliary clearance in the lung. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.P. Walentek, Signaling control of mucociliary epithelia: Stem cells, cell fates, and the plasticity of cell identity in development and disease. Cells Tissues Organs 211, 736–753 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.P. Satir, M. A. Sleigh, The physiology of cilia and mucociliary interactions. Annu. Rev. Physiol. 52, 137–155 (1990). [DOI] [PubMed] [Google Scholar]
- 7.E. R. Brooks, J. B. Wallingford, Multiciliated cells: A review. Curr. Biol. 24, R973–R982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. L. M. Hogan, C. E. Barkauskas, H. A. Chapman, J. A. Epstein, R. Jain, C. C. W. Hsia, L. Niklason, E. Calle, A. Le, S. H. Randell, J. Rock, M. Snitow, M. Krummel, B. R. Stripp, T. Vu, E. S. White, J. A. Whitsett, E. E. Morrisey, Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. T. Montoro, A. L. Haber, M. Biton, V. Vinarsky, B. Lin, S. E. Birket, F. Yuan, S. Chen, H. M. Leung, J. Villoria, N. Rogel, G. Burgin, A. M. Tsankov, A. Waghray, M. Slyper, J. Waldman, L. Nguyen, D. Dionne, O. Rozenblatt-Rosen, P. R. Tata, H. Mou, M. Shivaraju, H. Bihler, M. Mense, G. J. Tearney, S. M. Rowe, J. F. Engelhardt, A. Regev, J. Rajagopal, A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.P. Walentek, I. K. Quigley, What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopusto study cilia and mucociliary epithelia. Genesis 55, e23001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L. W. Plasschaert, R. Žilionis, R. Choo-Wing, V. Savova, J. Knehr, G. Roma, A. M. Klein, A. B. Jaffe, A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. C. Goldfarbmuren, N. D. Jackson, S. P. Sajuthi, N. Dyjack, K. S. Li, C. L. Rios, E. G. Plender, M. T. Montgomery, J. L. Everman, P. E. Bratcher, E. K. Vladar, M. A. Seibold, Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat. Commun. 11, 2485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.P. Walentek, Xenopus epidermal and endodermal epithelia as models for mucociliary epithelial evolution, disease, and metaplasia. Genesis 59, e23406 (2021). [DOI] [PubMed] [Google Scholar]
- 14.B. Mitchell, R. Jacobs, J. Li, S. Chien, C. Kintner, A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447, 97–101 (2007). [DOI] [PubMed] [Google Scholar]
- 15.G. A. Deblandre, D. A. Wettstein, N. Koyano-Nakagawa, C. Kintner, A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126, 4715–4728 (1999). [DOI] [PubMed] [Google Scholar]
- 16.P.-N. Tsao, M. Vasconcelos, K. I. Izvolsky, J. Qian, J. Lu, W. V. Cardoso, Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. Guirao, A. Meunier, S. Mortaud, A. Aguilar, J.-M. Corsi, L. Strehl, Y. Hirota, A. Desoeuvre, C. Boutin, Y.-G. Han, Z. Mirzadeh, H. Cremer, M. Montcouquiol, K. Sawamoto, N. Spassky, Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 12, 341–350 (2010). [DOI] [PubMed] [Google Scholar]
- 18.M. Haas, J. L. G. Vázquez, D. I. Sun, H. T. Tran, M. Brislinger, A. Tasca, O. Shomroni, K. Vleminckx, P. Walentek, ΔN-Tp63 mediates Wnt/β-catenin-induced inhibition of differentiation in basal stem cells of mucociliary epithelia. Cell Rep. 28, 3338–3352.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.J. A. Briggs, C. Weinreb, D. E. Wagner, S. Megason, L. Peshkin, M. W. Kirschner, A. M. Klein, The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Liao, L. Ma, Q. Guo, W. E, X. Fang, L. Yang, F. Ruan, J. Wang, P. Zhang, Z. Sun, H. Chen, Z. Lin, X. Wang, X. Wang, H. Sun, X. Fang, Y. Zhou, M. Chen, W. Shen, G. Guo, X. Han, Cell landscape of larval and adult Xenopus laevis at single-cell resolution. Nat. Commun. 13, 5142 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Green, The animal cap assay. Methods Mol. Biol. 127, 1–13 (1999). [DOI] [PubMed] [Google Scholar]
- 22.M. Blum, T. Ott, Xenopus: An undervalued model organism to study and model human genetic disease. Cells Tissues Organs 205, 303–313 (2018). [DOI] [PubMed] [Google Scholar]
- 23.P. D. Nieuwkoop, Normal Table of Xenopus Laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis (Garland Science, 1994). [Google Scholar]
- 24.M. Cibois, G. Luxardi, B. Chevalier, V. Thome, O. Mercey, L.-E. Zaragosi, P. Barbry, A. Pasini, B. Marcet, L. Kodjabachian, BMP signalling controls the construction of vertebrate mucociliary epithelia. Development 142, 2352–2363 (2015). [DOI] [PubMed] [Google Scholar]
- 25.E. Dubaissi, K. Rousseau, R. Lea, X. Soto, S. Nardeosingh, A. Schweickert, E. Amaya, D. J. Thornton, N. Papalopulu, A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development 141, 1514–1525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. Livigni, H. Peradziryi, A. A. Sharov, G. Chia, F. Hammachi, R. P. Migueles, W. Sukparangsi, S. Pernagallo, M. Bradley, J. Nichols, M. S. H. Ko, J. M. Brickman, A conserved Oct4/POUV-dependent network links adhesion and migration to progenitor maintenance. Curr. Biol. 23, 2233–2244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.G. E. Gentsch, T. Spruce, N. D. L. Owens, J. C. Smith, Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals. Nat. Commun. 10, 4269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. H. Levine, E. F. Simonds, S. C. Bendall, K. L. Davis, E.-A. D. Amir, M. D. Tadmor, O. Litvin, H. G. Fienberg, A. Jager, E. R. Zunder, R. Finck, A. L. Gedman, I. Radtke, J. R. Downing, D. Pe’er, G. P. Nolan, Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. D. Chalmers, K. Lachani, Y. Shin, V. Sherwood, K. W. Y. Cho, N. Papalopulu, Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech. Dev. 123, 702–718 (2006). [DOI] [PubMed] [Google Scholar]
- 30.G. Krasteva, U. Pfeil, M. Drab, W. Kummer, P. König, Caveolin-1 and -2 in airway epithelium: Expression and in situ association as detected by FRET-CLSM. Respir. Res. 7, 108 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. J. Aros, M. K. Paul, C. J. Pantoja, B. Bisht, P. Vijayaraj, J. M. Sandlin, L. K. Meneses, J. A. Tse, T. M. Rickabaugh, R. Damoiseaux, B. N. Gomperts, High-throughput drug screening identifies a potent wnt inhibitor that promotes airway basal stem cell homeostasis. Cell Rep. 30, 2055–2064.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. U. Hong, S. D. Reynolds, S. Watkins, E. Fuchs, B. R. Stripp, Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am. J. Pathol. 164, 577–588 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.V. Bergen, M. Lange, S. Peidli, F. A. Wolf, F. J. Theis, Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 34.G. La Manno, R. Soldatov, A. Zeisel, E. Braun, H. Hochgerner, V. Petukhov, K. Lidschreiber, M. E. Kastriti, P. Lönnerberg, A. Furlan, J. Fan, L. E. Borm, Z. Liu, D. van Bruggen, J. Guo, X. He, R. Barker, E. Sundström, G. Castelo-Branco, P. Cramer, I. Adameyko, S. Linnarsson, P. V. Kharchenko, RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.I. E. Royaux, S. M. Wall, L. P. Karniski, L. A. Everett, K. Suzuki, M. A. Knepper, E. D. Green, Pendrin, encoded by the Pendred syndrome gene, resides in the apical region of renal intercalated cells and mediates bicarbonate secretion. Proc. Natl. Acad. Sci. U.S.A. 98, 4221–4226 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.I. K. Quigley, J. L. Stubbs, C. Kintner, Specification of ion transport cells in the Xenopus larval skin. Development 138, 705–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Hu, Y.-R. Miao, L.-H. Jia, Q.-Y. Yu, Q. Zhang, A.-Y. Guo, AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.I. K. Quigley, C. Kintner, Rfx2 stabilizes Foxj1 binding at chromatin loops to enable multiciliated cell gene expression. PLOS Genet. 13, e1006538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Richard, L. Boullu, U. Herbach, A. Bonnafoux, V. Morin, E. Vallin, A. Guillemin, N. P. Gao, R. Gunawan, J. Cosette, O. Arnaud, J.-J. Kupiec, T. Espinasse, S. Gonin-Giraud, O. Gandrillon, Single-cell-based analysis highlights a surge in cell-to-cell molecular variability preceding irreversible commitment in a differentiation process. PLOS Biol. 14, e1002585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.G. S. Gulati, S. S. Sikandar, D. J. Wesche, A. Manjunath, A. Bharadwaj, M. J. Berger, F. Ilagan, A. H. Kuo, R. W. Hsieh, S. Cai, M. Zabala, F. A. Scheeren, N. A. Lobo, D. Qian, F. B. Yu, F. M. Dirbas, M. F. Clarke, A. M. Newman, Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Sedzinski, E. Hannezo, F. Tu, M. Biro, J. B. Wallingford, Emergence of an apical epithelial cell surface in vivo. Dev. Cell 36, 24–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. R. García, M. Deprez, K. Lebrigand, A. Cavard, A. Paquet, M.-J. Arguel, V. Magnone, M. Truchi, I. Caballero, S. Leroy, C.-H. Marquette, B. Marcet, P. Barbry, L.-E. Zaragosi, Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146, dev177428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.H. Y. Kim, T. R. Jackson, C. Stuckenholz, L. A. Davidson, Tissue mechanics drives regeneration of a mucociliated epidermis on the surface of Xenopus embryonic aggregates. Nat. Commun. 11, 665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. Dubaissi, N. Papalopulu, Embryonic frog epidermis: A model for the study of cell-cell interactions in the development of mucociliary disease. Dis. Model Mech. 4, 179–192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. M. Session, Y. Uno, T. Kwon, J. A. Chapman, A. Toyoda, S. Takahashi, A. Fukui, A. Hikosaka, A. Suzuki, M. Kondo, S. J. van Heeringen, I. Quigley, S. Heinz, H. Ogino, H. Ochi, U. Hellsten, J. B. Lyons, O. Simakov, N. Putnam, J. Stites, Y. Kuroki, T. Tanaka, T. Michiue, M. Watanabe, O. Bogdanovic, R. Lister, G. Georgiou, S. S. Paranjpe, I. van Kruijsbergen, S. Shu, J. Carlson, T. Kinoshita, Y. Ohta, S. Mawaribuchi, J. Jenkins, J. Grimwood, J. Schmutz, T. Mitros, S. V. Mozaffari, Y. Suzuki, Y. Haramoto, T. S. Yamamoto, C. Takagi, R. Heald, K. Miller, C. Haudenschild, J. Kitzman, T. Nakayama, Y. Izutsu, J. Robert, J. Fortriede, K. Burns, V. Lotay, K. Karimi, Y. Yasuoka, D. S. Dichmann, M. F. Flajnik, D. W. Houston, J. Shendure, L. DuPasquier, P. D. Vize, A. M. Zorn, M. Ito, E. M. Marcotte, J. B. Wallingford, Y. Ito, M. Asashima, N. Ueno, Y. Matsuda, G. J. C. Veenstra, A. Fujiyama, R. M. Harland, M. Taira, D. S. Rokhsar, Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. R. Rock, S. H. Randell, B. L. M. Hogan, Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545–556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Bilodeau, S. Shojaie, O. Goltsis, J. Wang, D. Luo, C. Ackerley, I. M. Rogers, B. Cox, M. Post, TP63 basal cells are indispensable during endoderm differentiation into proximal airway cells on acellular lung scaffolds. NPJ Regen Med. 6, 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. J. Arason, H. R. Jonsdottir, S. Halldorsson, B. E. Benediktsdottir, J. T. Bergthorsson, S. Ingthorsson, O. Baldursson, S. Sinha, T. Gudjonsson, M. K. Magnusson, deltaNp63 has a role in maintaining epithelial integrity in airway epithelium. PLOS ONE 9, e88683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.P. Walentek, S. Bogusch, T. Thumberger, P. Vick, E. Dubaissi, T. Beyer, M. Blum, A. Schweickert, A novel serotonin-secreting cell type regulates ciliary motility in the mucociliary epidermis of Xenopus tadpoles. Development 141, 1526–1533 (2014). [DOI] [PubMed] [Google Scholar]
- 50.C. Zechner, E. Nerli, C. Norden, Stochasticity and determinism in cell fate decisions. Development 147, dev181495 (2020). [DOI] [PubMed] [Google Scholar]
- 51.A. Raj, A. van Oudenaarden, Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C. A. Chacón-Martínez, J. Koester, S. A. Wickström, Signaling in the stem cell niche: Regulating cell fate, function and plasticity. Development 145, dev165399 (2018). [DOI] [PubMed] [Google Scholar]
- 53.C. S. Simon, A.-K. Hadjantonakis, C. Schröter, Making lineage decisions with biological noise: Lessons from the early mouse embryo. Wiley Interdiscip. Rev. Dev. Biol. 7, e319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. E. Werner, B. J. Mitchell, Understanding ciliated epithelia: The power of Xenopus. Genesis 50, 176–185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Ordovas-Montanes, D. F. Dwyer, S. K. Nyquist, K. M. Buchheit, M. Vukovic, C. Deb, M. H. Wadsworth 2nd, T. K. Hughes, S. W. Kazer, E. Yoshimoto, K. N. Cahill, N. Bhattacharyya, H. R. Katz, B. Berger, T. M. Laidlaw, J. A. Boyce, N. A. Barrett, A. K. Shalek, Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.F. A. V. Braga, G. Kar, M. Berg, O. A. Carpaij, K. Polanski, L. M. Simon, S. Brouwer, T. Gomes, L. Hesse, J. Jiang, E. S. Fasouli, M. Efremova, R. Vento-Tormo, C. Talavera-López, M. R. Jonker, K. Affleck, S. Palit, P. M. Strzelecka, H. V. Firth, K. T. Mahbubani, A. Cvejic, K. B. Meyer, K. Saeb-Parsy, M. Luinge, C.-A. Brandsma, W. Timens, I. Angelidis, M. Strunz, G. H. Koppelman, A. J. van Oosterhout, H. B. Schiller, F. J. Theis, M. van den Berge, M. C. Nawijn, S. A. Teichmann, A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 57.B. Treutlein, D. G. Brownfield, A. R. Wu, N. F. Neff, G. L. Mantalas, F. H. Espinoza, T. J. Desai, M. A. Krasnow, S. R. Quake, Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.C. Aztekin, T. W. Hiscock, J. C. Marioni, J. B. Gurdon, B. D. Simons, J. Jullien, Identification of a regeneration-organizing cell in the tail. Science 364, 653–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. L. Stubbs, L. Davidson, R. Keller, C. Kintner, Radial intercalation of ciliated cells during Xenopus skin development. Development 133, 2507–2515 (2006). [DOI] [PubMed] [Google Scholar]
- 60.A. Tasca, M. Helmstädter, M. M. Brislinger, M. Haas, B. Mitchell, P. Walentek, Notch signaling induces either apoptosis or cell fate change in multiciliated cells during mucociliary tissue remodeling. Dev. Cell 56, 525–539.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.C. Aztekin, T. W. Hiscock, J. B. Gurdon, J. Jullien, J. C. Marioni, B. D. Simons, Secreted inhibitors drive the loss of regeneration competence in Xenopus limbs. Development 148, dev199158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.K. S. Dingwell, J. C. Smith, Dissecting and culturing animal cap explants. Cold Spring Harb. Protoc. 2018, pdb.prot097329 (2018). [DOI] [PubMed] [Google Scholar]
- 63.G. X. Y. Zheng, J. M. Terry, P. Belgrader, P. Ryvkin, Z. W. Bent, R. Wilson, S. B. Ziraldo, T. D. Wheeler, G. P. McDermott, J. Zhu, M. T. Gregory, J. Shuga, L. Montesclaros, J. G. Underwood, D. A. Masquelier, S. Y. Nishimura, M. Schnall-Levin, P. W. Wyatt, C. M. Hindson, R. Bharadwaj, A. Wong, K. D. Ness, L. W. Beppu, H. J. Deeg, C. McFarland, K. R. Loeb, W. J. Valente, N. G. Ericson, E. A. Stevens, J. P. Radich, T. S. Mikkelsen, B. J. Hindson, J. H. Bielas, Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. Lukoseviciute, R. Lea, S. Ishibashi, E. Amaya, Whole-mount in situ hybridization and a genotyping method on single Xenopus embryos. Basic Sci. Methods Clin. Researchers , 41–56 (2017). [Google Scholar]
- 65.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Frankish, M. Diekhans, A.-M. Ferreira, R. Johnson, I. Jungreis, J. Loveland, J. M. Mudge, C. Sisu, J. Wright, J. Armstrong, I. Barnes, A. Berry, A. Bignell, S. C. Sala, J. Chrast, F. Cunningham, T. Di Domenico, S. Donaldson, I. T. Fiddes, C. G. Girón, J. M. Gonzalez, T. Grego, M. Hardy, T. Hourlier, T. Hunt, O. G. Izuogu, J. Lagarde, F. J. Martin, L. Martínez, S. Mohanan, P. Muir, F. C. P. Navarro, A. Parker, B. Pei, F. Pozo, M. Ruffier, B. M. Schmitt, E. Stapleton, M.-M. Suner, I. Sycheva, B. Uszczynska-Ratajczak, J. Xu, A. Yates, D. Zerbino, Y. Zhang, B. Aken, J. S. Choudhary, M. Gerstein, R. Guigó, T. J. P. Hubbard, M. Kellis, B. Paten, A. Reymond, M. L. Tress, P. Flicek, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.G. Marçais, C. Kingsford, A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. M. Rodriguez, J. Rodriguez-Rivas, T. Di Domenico, J. Vázquez, A. Valencia, M. L. Tress, APPRIS 2017: Principal isoforms for multiple gene sets. Nucleic Acids Res. 46, D213–D217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. D. Fortriede, T. J. Pells, S. Chu, P. Chaturvedi, D. Wang, M. E. Fisher, C. James-Zorn, Y. Wang, M. J. Nenni, K. A. Burns, V. S. Lotay, V. G. Ponferrada, K. Karimi, A. M. Zorn, P. D. Vize, Xenbase: Deep integration of GEO & SRA RNA-seq and ChIP-seq data in a model organism database. Nucleic Acids Res. 48, D776–D782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. Parekh, C. Ziegenhain, B. Vieth, W. Enard, I. Hellmann, zUMIs—A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, giy059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.A. T. L. Lun, S. Riesenfeld, T. Andrews, T. P. Dao, T. Gomes; participants in the 1st Human Cell Atlas Jamboree, J. C. Marioni, EmptyDrops: Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.S. L. Wolock, R. Lopez, A. M. Klein, Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.F. A. Wolf, P. Angerer, F. J. Theis, SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.L. McInnes, J. Healy, N. Saul, L. Großberger, UMAP: Uniform manifold approximation and projection. J. Open Source Softw.. 3, 861 (2018). [Google Scholar]
- 76.D. van Dijk, R. Sharma, J. Nainys, K. Yim, P. Kathail, A. J. Carr, C. Burdziak, K. R. Moon, C. L. Chaffer, D. Pattabiraman, B. Bierie, L. Mazutis, G. Wolf, S. Krishnaswamy, D. Pe’er, Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.M. Setty, V. Kiseliovas, J. Levine, A. Gayoso, L. Mazutis, D. Pe’er, Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 ((2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. Nowotschin, M. Setty, Y.-Y. Kuo, V. Liu, V. Garg, R. Sharma, C. S. Simon, N. Saiz, R. Gardner, S. C. Boutet, D. M. Church, P. A. Hoodless, A.-K. Hadjantonakis, D. Pe’er, The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. Shahsavari, A. Munteanu, I. Mohorianu, ClustAssess: Tools for assessing the robustness of single-cell clustering. bioRxiv 2022.01.31.478592 [Preprint]. 2 February 2022. 10.1101/2022.01.31.478592. [DOI]
- 80.G. Yu, L.-G. Wang, Y. Han, Q.-Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.A. Santos, R. Wernersson, L. J. Jensen, Cyclebase 3.0: A multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res. 43, D1140–D1144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Notes S1 to S4
Figs. S1 to S15
Legends for tables S1 to S12
Tables S1 to S12