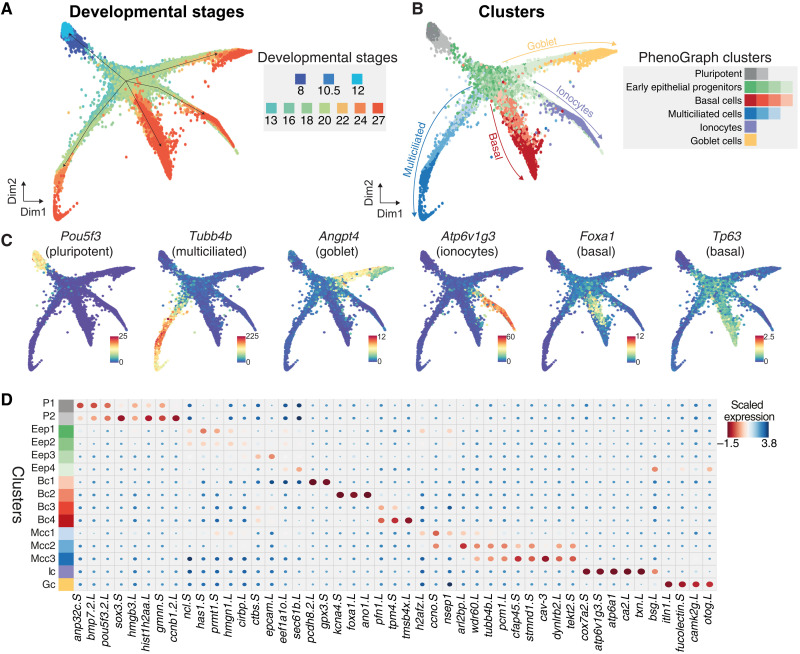
Fig. 2. Joint embedding of continuous MCE developmental manifold and cell-type differentiation.
(A) High-density force-directed k-nearest neighbor (knn) graph visualization of single cells, colored by developmental stages. (B) PhenoGraph clustering of single cells over continuous MCE manifold, colored by different cell types and cell states (subclusters). The arrows indicate the differentiation of progenitors into specific cell types. (C) Expression patterns of marker genes for MCE cell types overlaid on the knn graph. The color bars indicated scaled imputed expression levels. (D) Dot plot of marker genes across different MCE clusters. The color represents maximum normalized mean marker gene expression across each cluster, and the size indicates the proportion of positive cells relative to the entire dataset.