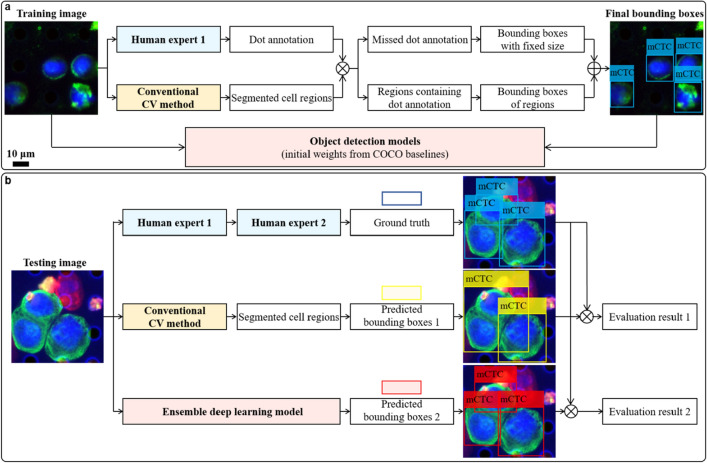
Figure 5.
Cell detection via deep learning. (a) Training pipeline. An experienced pathologist annotates the cells of interest in training images with dots and simultaneously these images are processed by a conventional computer vision (CV) method to segment cell regions. Results from both methods are cross validated by matching annotation dots and segmentation regions. Any region containing annotation dots is used to generate a bounding box and paired with the annotation label. For dots which do not lie in any region, a bounding box centered at each of them is generated with the size of empirical cell diameter. Then, training images and their corresponding bounding boxes with class labels are used to train a generic object detection deep learning model. Here, transfer learning is adopted by using weights pretrained on the COCO benchmark dataset. (b) Testing pipeline. The unseen testing images are analyzed in three ways. First, the same experienced pathologist screens testing images by annotating the cells of interest with bounding boxes, which are sequentially double checked by another computational pathology researcher to make sure there is no oversight or mislabeling. This result is taken as ground truth. In parallel, testing images are segmented by the conventional CV method and then the prediction boxes with labels are generated from segmented regions. Finally, they are sent to our well-trained cell detection model and the predicted bounding boxes can be directly generated. Comparing results from the latter two methods with the ground truth, we find our trained deep learning model outperforms the conventional CV method.