Mutations in rhodopsin are the major cause of autosomal dominant retinitis pigmentosa. Zhao et al. demonstrated that PERK signaling plays key roles in maintaining rhodopsin homeostasis through attenuating IRE1-induced autophagy, and upregulation of PERK prevents autophagy and suppresses retinal degeneration.
Abstract
Chronic endoplasmic reticulum (ER) stress is the underlying cause of many degenerative diseases, including autosomal dominant retinitis pigmentosa (adRP). In adRP, mutant rhodopsins accumulate and cause ER stress. This destabilizes wild-type rhodopsin and triggers photoreceptor cell degeneration. To reveal the mechanisms by which these mutant rhodopsins exert their dominant-negative effects, we established an in vivo fluorescence reporter system to monitor mutant and wild-type rhodopsin in Drosophila. By performing a genome-wide genetic screen, we found that PERK signaling plays a key role in maintaining rhodopsin homeostasis by attenuating IRE1 activities. Degradation of wild-type rhodopsin is mediated by selective autophagy of ER, which is induced by uncontrolled IRE1/XBP1 signaling and insufficient proteasome activities. Moreover, upregulation of PERK signaling prevents autophagy and suppresses retinal degeneration in the adRP model. These findings establish a pathological role for autophagy in this neurodegenerative condition and indicate that promoting PERK activity could be used to treat ER stress-related neuropathies, including adRP.
Introduction
Defects in protein folding are a common cellular event, typically resulting from genetic mutations, translational errors, or a range of cellular stresses. Thus, maintaining an intact proteasome and cellular function requires continuous removal of misfolded proteins (Kurtishi et al., 2019). Eukaryotic cells are equipped with a number of physiological mechanisms to ensure proteins are correctly folded and to degrade misfolded proteins, but a prolonged imbalance between the generation of misfolded proteins and quality control mechanisms can disrupt cellular function. This underlies many diseases, including neurodegenerative disorders (Klaips et al., 2018). In eukaryotic cells, the ER is an intracellular organelle central to the synthesis of secretory and membrane proteins (Sano and Reed, 2013). When cells experience stress (e.g., oxidative stress or aging), the accumulation of misfolded proteins results in a loss of proteostasis. These misfolded proteins accumulate in the ER resulting in the activation of the unfolded protein response (UPR). The UPR is a cellular homeostatic mechanism that reduces ER stress by promoting the degradation of misfolded proteins and slowing the synthesis of new proteins (Hetz et al., 2020; Walter and Ron, 2011).
The UPR is controlled by three ER-resident transmembrane proteins, inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase RNA-like ER kinase (PERK; Walter and Ron, 2011). Upon ER stress, PERK oligomerizes, leading to phosphorylation of eukaryotic initiation factor 2α (eIF2α). Phosphorylated eIF2α binds and inhibits the guanine nucleotide exchange factor, eIF2B, thereby attenuating eIF2-mediated protein synthesis (Adomavicius et al., 2019; Kenner et al., 2019). In contrast with the global repression of translation, eIF2α phosphorylation also activates the stress-responsive transcription factors, ATF4 and Xrp1, through selectively enhanced translation (Brown et al., 2021; Harding et al., 1999; Harding et al., 2003). Xrp1 is a newly discovered transcription factor induced downstream of PERK in Drosophila (Brown et al., 2021). In addition, UPR signaling activates the IRE1 nuclease, which targets and splices mRNA encoding the transcription factor X-box-binding protein 1 (XBP1), thereby activating it. Activated XBP1 then upregulates genes involved in ER protein folding, as well as genes promoting the degradation of misfolded proteins (Calfon et al., 2002; Cox et al., 1993; Haze et al., 1999; Yoshida et al., 2001). Transcriptional targets of XBP1 and ATF6 overlap significantly; the latter undergoes stress-induced intramembrane proteolytic processing and translocates to the nucleus (Mori et al., 1993; Shoulders et al., 2013). The three UPR pathways, in particular the IRE1 and PERK branches, have different activating states, and unequal or contradictory effects on cellular pathophysiology, depending on the disease and physiological context (Chang et al., 2018; Lin et al., 2007; Zhu et al., 2019). Consistent with their different activating states, the inhibition of translation by PERK attenuates IRE1 activation following a prolonged UPR state. However, the mechanisms by which one UPR branch affects another and the physiological significance of this regulation are not understood (Chang et al., 2018; Lin et al., 2007).
Autosomal dominant retinitis pigmentosa (adRP), the most common form of retinal degeneration, is most often caused by dominant mutations in the rhodopsin gene (Rho). Resulting mutant G protein-coupled receptors (GPCR) are misfolded and accumulate in the ER (Athanasiou et al., 2018; Hartong et al., 2006; Mendes et al., 2005). The substitution of proline 23 by histidine (RHOP23H), the most common adRP-associated mutation, results in rhodopsin improper folding, retention in the ER, activation of the UPR, and ultimately photoreceptor degeneration (Dryja et al., 1990; Lin et al., 2007). Interestingly, this mutated opsin exerts a dominant negative effect on wild-type RHO, as co-expression of wild-type RHO and RHOP23H results in: (1) mislocalized wild-type RHO, (2) formation of inclusions that contain wild-type RHO, and (3) enhanced proteasome-mediated degradation of wild-type RHO (Mendes and Cheetham, 2008; Rajan and Kopito, 2005; Saliba et al., 2002). In a RhoP23H knock-in mouse model, levels of wild-type RHO are decreased, and heterozygous animals exhibit retinal degeneration in the rod outer segment (Sakami et al., 2011). Similarly, Drosophila carrying a heterozygous mutation in the major rhodopsin, Rh1 (ninaEG69D), which is encoded by the ninaE locus, exhibits low levels of both mutated and wild-type Rh1 (Colley et al., 1995). Compounds that reduce the dominant-negative effects of the RHOP23H opsin alleviate cell death, suggesting that the interruption of opsin homeostasis by dominant RHO is involved in adRP pathology (Mendes and Cheetham, 2008). Although it is clear that misfolded rhodopsin dominant negatively affects the wild-type protein, the mechanisms and physiological role have not been identified.
IRE1 and ATF6 can both upregulate the expression of chaperones involved in RHO folding, as well as ER-associated degradation (ERAD) components that specifically remove and degrade misfolded proteins via the ubiquitin–proteasome system (UPS), thereby leading to the degradation of misfolded RHO while sparing the wild-type version (Chiang et al., 2015; Chiang et al., 2012b; Lin et al., 2007; Ryoo et al., 2007; Shoulders et al., 2013). In addition to ERAD, the autophagy pathway, which is a second cellular quality control mechanism for clearing damaged proteins, is actively involved in regulating turnover of ER proteins and the ER itself (Khaminets et al., 2015). Induction of autophagy is observed in RhoP23H mice, leading to proteasome insufficiency and increased retinal degeneration (Qiu et al., 2019; Yao et al., 2018). It has been suggested that the levels of autophagy increase as a result of the loss of Atf6 (Lee et al., 2021). Recently in a Drosophila model of Parkinson’s disease, overexpression of IRE1 was shown to induce autophagy and triggers neuronal cell death in an XBP1-independent manner (Yan et al., 2019). By contrast to the selective reduction of mutant opsin through ATF6 or IRE1 signaling, the PERK pathway shows no such specificity, reducing both wild-type and mutant RHO protein levels. This suggests that PERK is involved in the non-selectively removal of ER proteins (Chiang et al., 2012a). It remains unclear how the UPR induces autophagy, and whether the selective autophagy of ER is responsible for maintaining cellular protein homeostasis.
In the present study, we established an in vivo model in Drosophila to study the dominant effects of misfolded rhodopsin on the wild-type protein. In this model, we changed proline 37 to histidine (Rh1P37H is equivalent to mammalian RHOP23H) and tagged this mutant opsin with GFP. We also tagged wild-type Rh1 with RFP and co-expressed these proteins in photoreceptor cells (Galy et al., 2005; Griciuc et al., 2010). Using this newly developed system, we conducted a forward genetic screen to identify genes that enhance the dominant effects of Rh1P37H. We found that the PERK pathway played a key role in maintaining levels of wild-type Rh1 and in sustaining photoreceptor cell function and integrity in Rh1P37H-expressing cells. This effect was independent of ATF4. In animals lacking the PERK pathway, IRE1/XBP1 signaling was over-activated, leading to massive autophagy and degradation of wild-type rhodopsin. Finally, in a fly model of adRP, induction of PERK signaling prevented the induction of autophagy and thus suppressed retinal degeneration.
Results
Rhodopsin homeostasis is disrupted by mutations in perk and eIF2Bα in a dominant Rh1 mutant fly model
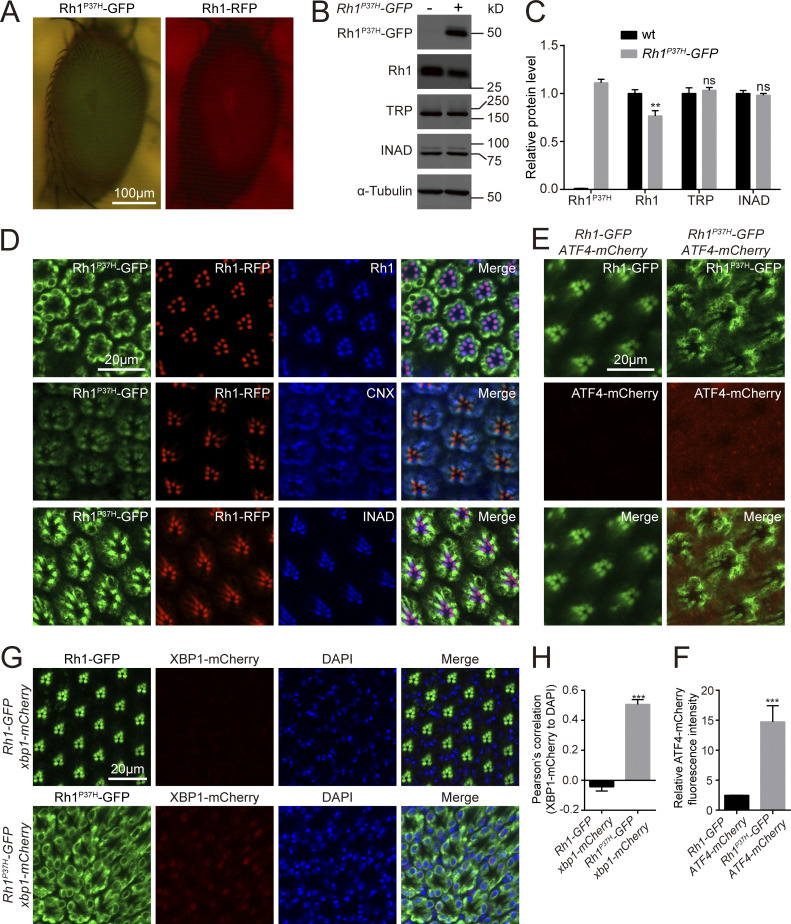
To study the genetic interactions between misfolded and wild-type versions of rhodopsin in vivo, we modified the previously established Drosophila model of adRP by co-expressing GFP-tagged Rh1P37H and RFP-tagged wild-type Rh1 using the endogenous ninaE promotor (ninaE, neither inactivation nor afterpotential E; Fig. 1 A; Galy et al., 2005; Griciuc et al., 2010; O’Tousa et al., 1985; Zuker et al., 1985). Consistent with previous reports, Rh1P37H-GFP accumulated exclusively in the ER, co-localizing with the ER marker CNX (calnexin). By contrast, wild-type Rh1-RFP localized to the rhabdomeres with endogenous Rh1 and INAD (Fig. 1 D; Galy et al., 2005). Moreover, Rh1P37H-GFP induced ER stress, as both ATF4-mCherry and XBP1-mCherry (two independent reporters of ER stress) were expressed and activated in Rh1P37H-GFP retinas, but not in retinas expressing wild-type Rh1-GFP (Kang and Ryoo, 2009; Xu et al., 2020; Fig. 1, E–H). Importantly, expression of misfolded Rh1P37H-GFP resulted in less wild-type Rh1 but did not affect levels of endogenous TRP and INAD. This indicates that disease-causing rhodopsin mutations mildly impaired rhodopsin homeostasis (Fig. 1, B and C).
Figure 1.
Establishment of a Rh1P37H-GFP-based Drosophila model of autosomal dominant retinitis pigmentosa (adRP). (A) Representative images of compound eyes expressing Rh1P37H-GFP and Rh1-RFP. Scale bar, 100 μm. (B) Western blot revealed that expression of Rh1P37H-GFP reduced the endogenous protein levels of wild-type Rh1. Anti-Rh1 antibodies failed to recognize c-terminal tagged Rh1 (Rh1-GFP/RFP). 1-d-old flies raised under 12-h-light–12-h-dark cycles were used. α-tubulin was used as a loading control. (C) Quantification of relative levels of endogenous Rh1, TRP, and INAD from B. Error bars indicate SEM (n = 3); ns, not significant; **P < 0.01 (two-way ANOVA, Sidak’s multiple comparisons test). (D) Tangential views of retina from ∼1-d-old Rh1-RFP; Rh1P37H-GFP flies labeled using anti-Rh1, anti-CNX (calnexin), and anti-INAD antibodies (blue). GFP fluorescence of Rh1P37H-GFP (green) and RFP fluorescence of Rh1-RFP (red) were directly observed. Scale bar, 20 μm. (E) Retinas of adult Rh1-GFP and Rh1P37H-GFP flies expressing ATF4-mCherry.1-d-old flies raised under 12-h-light–12-h-dark cycles were used. Scale bar, 20 μm. (F) Quantification of relative mCherry fluorescence intensity showed that the ER stress reporter ATF4-mCherry (E) was activated by Rh1P37H-GFP but not by wild-type Rh1-GFP. Error bars indicate SEM (n = 3); ***P < 0.001 (Student’s unpaired t test). (G) Retinas of adult Rh1-GFP and Rh1P37H-GFP flies expressing xbp1-mCherry. 1-d-old flies raised under 12-h-light–12-h-dark cycles were used. Scale bar, 20 μm. (H) Quantification of the co-localization between XBP1-mCherry and DAPI in flies that express Rh1-GFP or Rh1P37H-GFP. Error bars indicate SEM (n = 3); ***P < 0.001 (Student’s unpaired t test). Source data are available for this figure: SourceData F1.
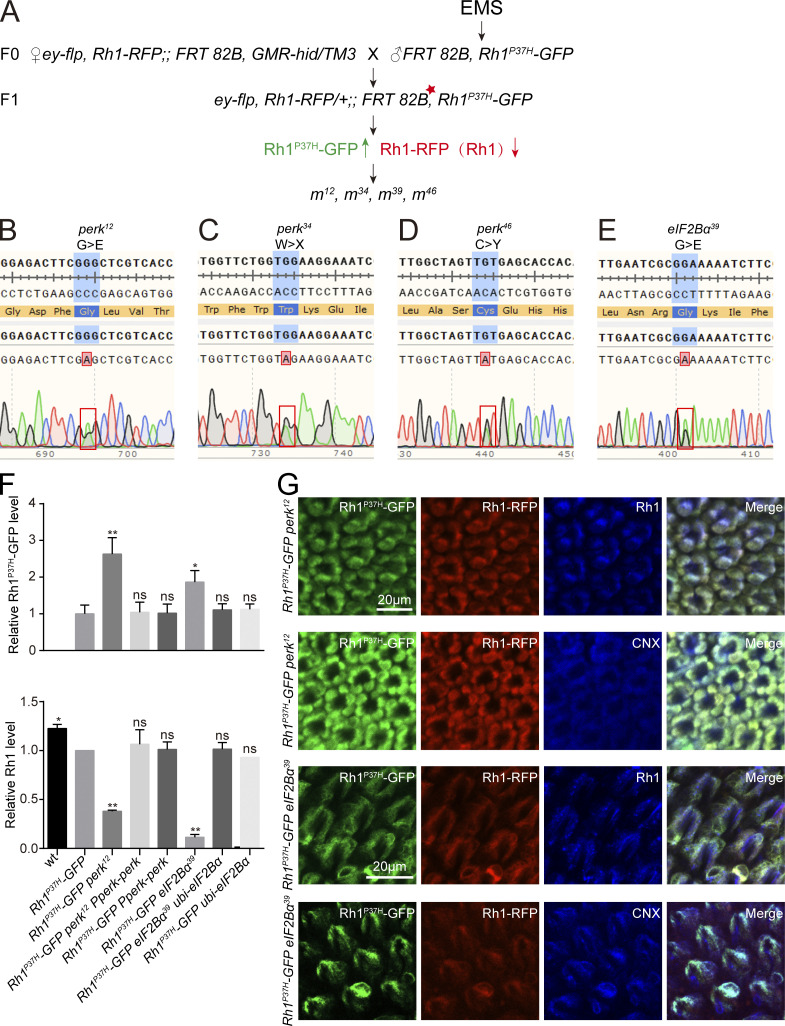
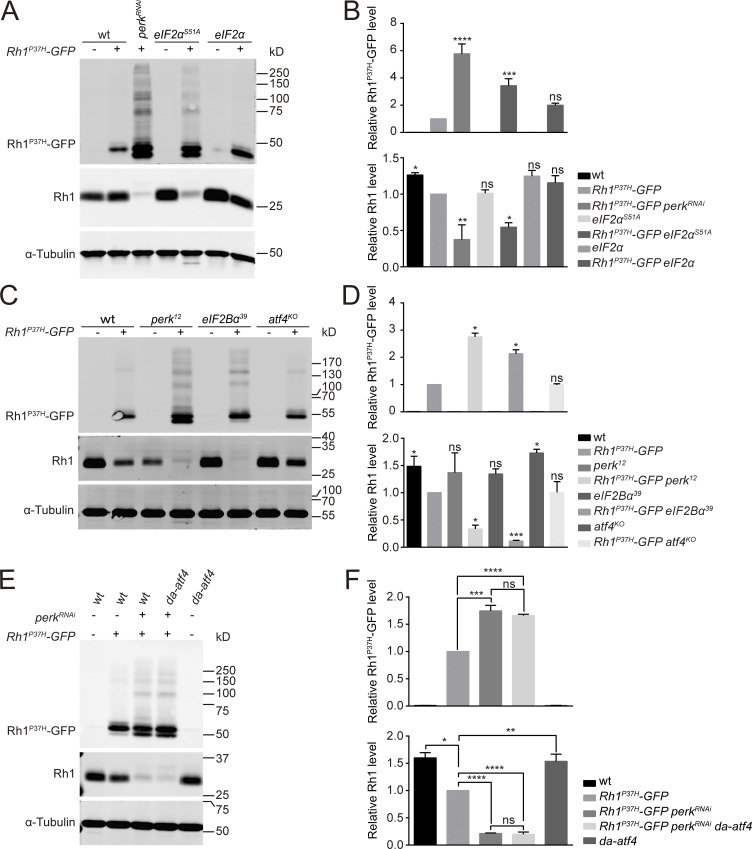
Combining this Rh1P37H-GFP/Rh1-RFP reporters with the “ey-flp/hid” system, which generates flies in which EMS-induced mutations are homozygous in the eye but heterozygous in the rest of the animal, we performed EMS mutagenesis and screened chromosomes 2 and 3 (including 2L, 2R, 3L, and 3R) for mutants in which Rh1P37H-GFP further disrupted the homeostasis of wild-type Rh1 (Fig. S1 A; Xiong et al., 2020; Zhao et al., 2018). For each chromosome arm we screened ∼100,000 flies, isolating 4 alleles in which RFP but not GFP fluorescence was reduced. These four alleles belonged to two complementation groups, both located on the right arm of chromosome 3 (3R). Using deficiency mapping and genomic DNA sequencing, we found that one complementation group localized to the perk locus (perk12, perk34, perk46), and the other localized to the eIF2Bα gene (eIF2Bα39; Fig. 2, A and B). These four alleles contain single-nucleotide changes within the coding region causing missense mutations in PERK and eIF2Bα proteins (Fig. S1, B–E). Flies heterozygous for any of these perk or eIF2Bα alleles did not exhibit a phenotype, regardless of whether they express Rh1P37H-GFP or not, suggesting that these are loss of function mutations. We confirmed via Western blotting that levels of endogenous wild-type Rh1 were greatly reduced in perk12 and eIF2Bα39 mutant animals, whereas Rh1P37H-GFP accumulated (Fig. 2 C and Fig. S1 F). Importantly, the lower MW Rh1P37H-GFP bands in perk12 and eIF2Bα39 mutant extracts are non-glycosylated versions of the protein, suggesting that ER function was impaired. Moreover, expression of wild-type PERK or eIF2Bα in perk12 or eIF2Bα39 mutants, respectively, restored levels of endogenous Rh1 and reduced levels of Rh1P37H-GFP to control levels (Fig. 2 C and Fig. S1 F).
Figure S1.
Genetic screen reveals that perk and eIF2Bα are involved in Rh1 homeostasis in a model of adRP. (A) EMS screening strategy to identify factors that regulate Rh1 homeostasis when Rh1P37H is expressed. Screening the right arm of the third chromosome (3R) is used as an example. The FRT82B Rh1P37H-GFP flies were isogenized, and male flies were fed 25 mM EMS (Sigma-Aldrich) in 2% sucrose for 8 h, followed by mating to ey-flp Rh1-RFP; FRT82B GMR-hid CL/TM3 flies. Approximately 100,000 F1 progeny with homozygous mutant eyes were examined for fluorescence of GFP-tagged Rh1P37H and RFP-tagged wild-type Rh1. (B–E) Mutations associated with the perk12, perk34, perk46, and eIF2Bα39 alleles. (F) Quantification of protein levels of Rh1P37H-GFP and endogenous Rh1 shown in Fig. 2 C. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01 (one-way ANOVA, Sidak’s multiple comparisons test). (G) Staining perk12, and eIF2Bα39 retina expressing Rh1P37H-GFP and Rh1-RFP against CNX (blue, ER marker) and wild-type Rh1 (blue). GFP fluorescence of Rh1P37H-GFP (green) and RFP fluorescence of Rh1-RFP (red) were directly observed. Scale bar, 20 μm.
Figure 2.
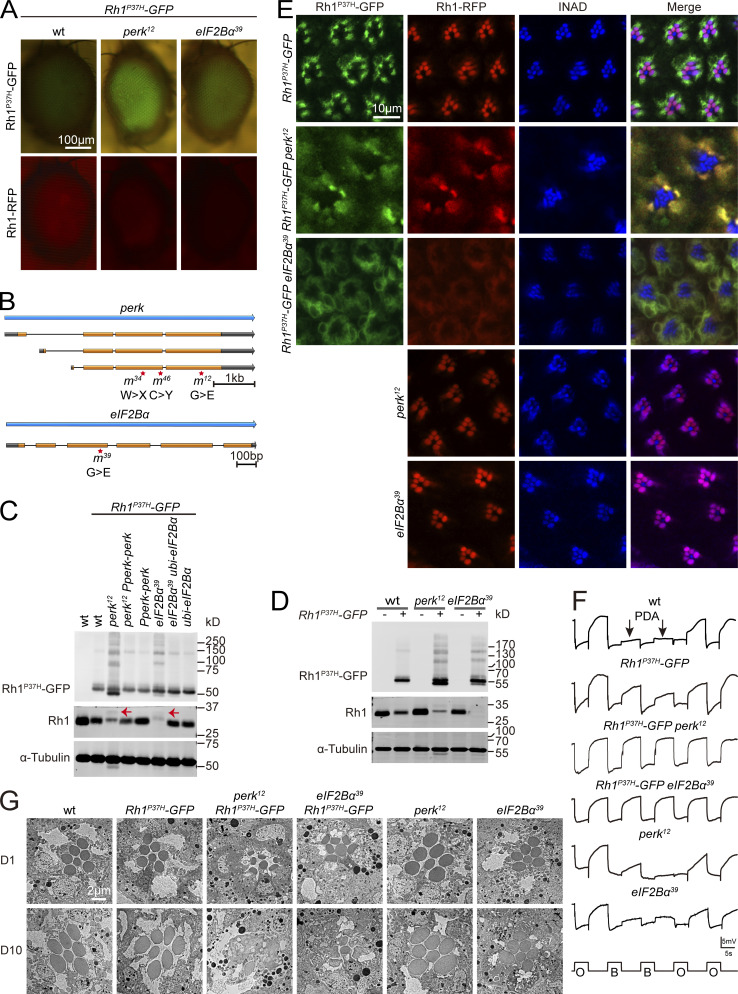
Rhodopsin homeostasis is disrupted by mutations in perk or eIF2Bα in the adRP model. (A) Isolation of perk12 and eIF2Bα39 mutants via a forward genetic screen. Rh1-RFP and Rh1P37H–GFP fluorescence were detected using a Stereo Fluorescence Microscope. Images from wild-type (ey-flp Rh1-RFP; Rh1P37H-GFP), perk12 (ey-flp Rh1-RFP; FRT82B Rh1P37H-GFP perk12/FRT82B GMR-hid CL), and eIF2Bα39 (ey-flp Rh1-RFP; FRT82B Rh1P37H-GFP eIF2Bα39/FRT82B GMR-hid CL) flies are shown. Scale bar, 100 μm. (B) The perk and eIF2Bα loci and mutations associated with the perk12, perk34, perk46, and eIF2Bα39 alleles. (C) Western blot of heads dissected from wild-type, perk12, and eIF2Bα39 flies expressing Rh1P37H-GFP. Levels of GFP and Rh1 are shown. Expressing PERK via the endogenous perk promoter (Pperk-perk) and eIF2Bα under a ubiquitin promoter (ubi-eIF2Bα) rescued the phenotypes. 1-d-old flies were used, and α-tubulin was used as a loading control. Bands of Rh1 with increased molecular weight (MW) in Rh1P37H-GFP perk12 and Rh1P37H-GFP eIF2Bα39 mutant flies are indicated by red arrows. (D) Western blot analysis of Rh1 in homozygous perk12 (ey-flp Rh1-RFP; FRT82B perk12/FRT82B GMR-hid CL) and eIF2Bα39 (ey-flp Rh1-RFP; FRT82B eIF2Bα39/FRT82B GMR-hid CL) mutants without Rh1P37H expression. (E) Tangential views of wild-type, perk12, and eIF2Bα39 retina expressing Rh1P37H-GFP and Rh1-RFP or homozygous perk12 (ey-flp Rh1-RFP; FRT82B perk12/FRT82B GMR-hid CL) and eIF2Bα39 (ey-flp Rh1-RFP; FRT82B eIF2Bα39/FRT82B GMR-hid CL) mutants without Rh1P37H expression labeled for INAD (blue, a rhabdomere marker). GFP fluorescence of Rh1P37H-GFP (green) and RFP fluorescence of Rh1-RFP (red) were directly observed. Scale bar, 10 μm. (F) ERG recordings of wt (Rh1-GFP) and Rh1P37H-GFP (ey-flp Rh1-RFP; Rh1P37H-GFP) flies showed that a PDA was induced by blue light (arrows). The PDA was eliminated in perk12 (ey-flp Rh1-RFP; FRT82B Rh1P37H-GFP perk12/FRT82B GMR-hid CL), and eIF2Bα39 (ey-flp Rh1-RFP; FRT82B Rh1P37H-GFP eIF2Bα39/FRT82B GMR-hid CL) flies. ERG recordings of perk12 and eIF2Bα39 mutants without Rh1P37H expression showed normal PDAs. 1-d-old flies were exposed to 5-s pulses of orange (O) or blue (B) light as indicated. At least 10 flies of each genotype were tested. (G) TEM images of eye tangential sections from 1-d-old and 10-d-old flies. Genotypes are indicated. Scale bar, 2 μm. All flies were in white eye background and raised under 12 h light/12 h dark cycles. Source data are available for this figure: SourceData F2.
During biosynthesis, Rh1 is transiently glycosylated in the ER. This modification is gradually removed as Rh1 is transported from the ER to the rhabdomere (Rosenbaum et al., 2014). A band of Rh1 with an increased molecular weight (MW) was observed in Rh1P37H-GFP perk12 and Rh1P37H-GFP eIF2Bα39 mutant flies, indicating a defective maturation process for wild-type rhodopsin (Fig. 2 C). We then examined the localization of wild-type Rh1 and Rh1P37H-GFP in perk12 and eIF2Bα39 mutants. In these mutant retinas, both wild-type Rh1-RFP and endogenous Rh1 colocalized with Rh1P37H-GFP in the ER, whereas INAD still localized to the rhabdomeres. This indicates that trafficking of wild-type Rh1 was disrupted by perk and eIF2Bα mutations with misfolded Rh1P37H expression (Fig. 2 E and Fig. S1 G). We next asked if mutations in perk and eIF2Bα affected the biosynthesis and trafficking of rhodopsin. We found that Rh1 levels and localization were normal in both perk12 and eIF2Bα39 mutant photoreceptor cells (without Rh1P37H expression; Fig. 2, D and E). These results indicate that PERK and eIF2Bα help maintain the homeostasis of wild-type rhodopsin when misfolded form is present.
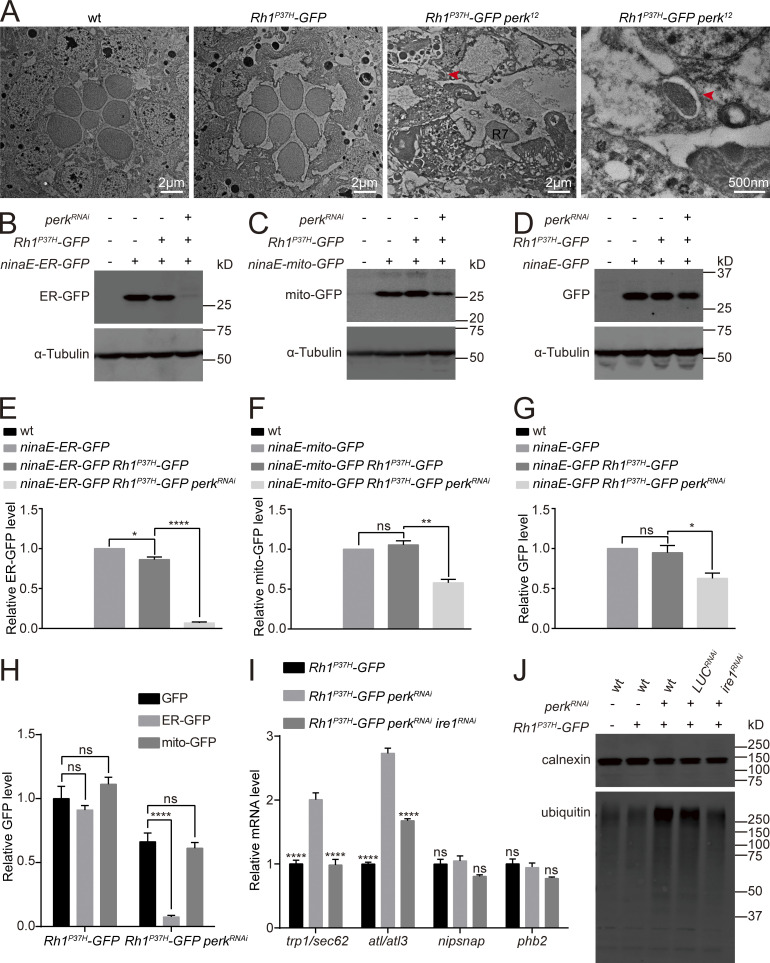
Since Rh1 is essential for photoreceptor function, we asked whether phototransduction was disrupted in perk and eIF2Bα mutants. ERG (electroretinogram) recordings measure the summed light responses of all retinal cells. In flies with normal levels of functional rhodopsin, a prolonged depolarization afterpotential (PDA) is induced upon exposure to blue light (Fig. 2 F; Wang and Montell, 2007). Flies expressing Rh1P37H exhibited a normal PDA, consistent with a slight reduction in wild-type Rh1 levels. However, no PDAs were detected in perk12 and eIF2Bα39 mutants expressing Rh1P37H, consistent with the large reduction in wild-type Rh1 levels (Fig. 2 F). In contrast, perk12 and eIF2Bα39 mutants that lacked Rh1P37H expression exhibited normal PDAs when exposed to blue light (Fig. 2 F). As disruptions in rhodopsin homeostasis are associated with the progression of retinal degeneration in adRP diseases, we next asked if disrupting PERK and eIF2Bα aggravates the severity of retinal degeneration in the Rh1P37H-GFP model. We used TEM to first assess young (1-d-old) and aged (10-d-old) wild-type, Rh1P37H-GFP, perk12, and eIF2Bα39 flies. For each age and genotype, seven intact rhabdomeres were consistently detected in each ommatidia (Fig. 2 G). In the Rh1P37H-GFP background, young perk12 and eIF2Bα39 mutants exhibited normal retinal morphology with all 7 rhabdomeres, although the rhabdomeres were smaller. However, aged flies of these genotypes (perk12 or eIF2Bα39 mutations in the Rh1P37H-GFP background) exhibited severe retinal degeneration with prominent vacuoles and loss of rhabdomeres (Fig. 2 G). Therefore, we conclude that PERK and eIF2Bα are required for photoreceptor survival in the context of Rh1P37H-induced ER stress.
The PERK pathway is the major UPR axis maintaining rhodopsin homeostasis independent of ATF4
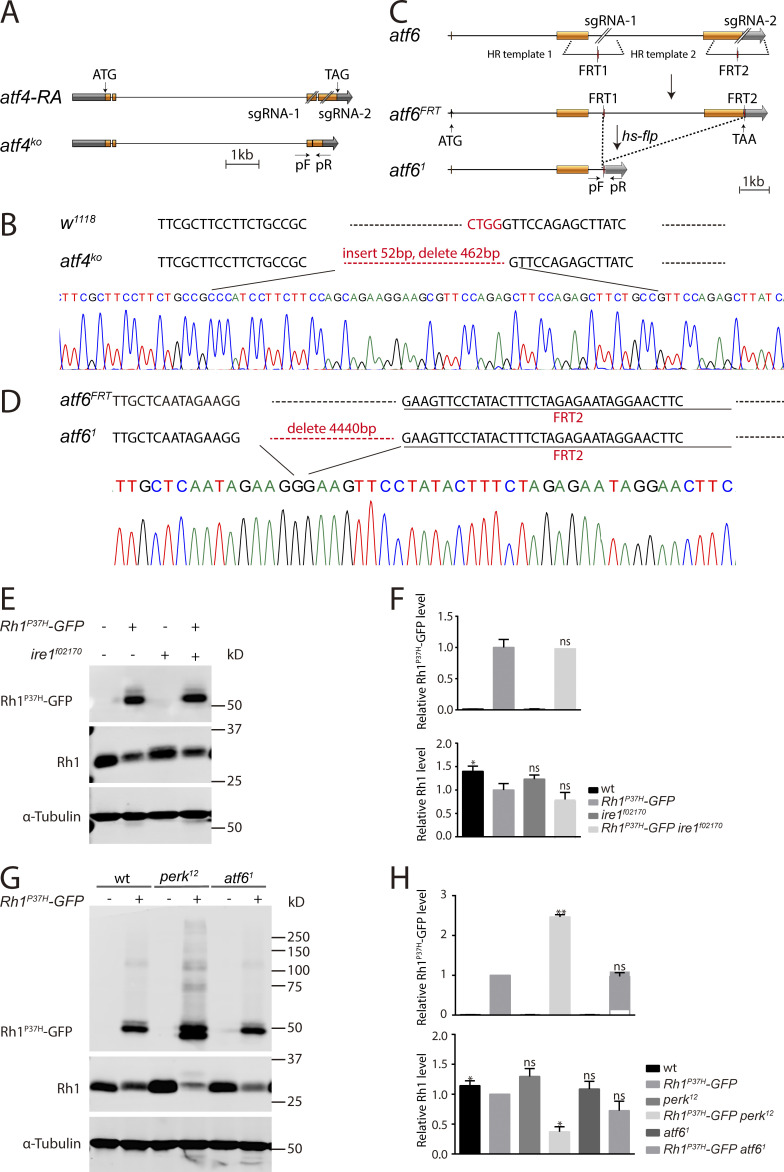
Misfolded rhodopsin causes ER stress and activates three UPR pathways, namely, the PERK, IRE1, and ATF6 pathways (Harding et al., 2000b; Liu et al., 2000; Shamu and Walter, 1996; Shen et al., 2002). Once activated, PERK phosphorylates eIF2α and promotes its inhibitory binding to the nucleotide exchange factor eIF2B to regulate translation initiation. Therefore, mutations in either the perk or eIF2Bα genes would impair the PERK/eIF2α branch of the UPR response. Since our screen did not identify genes involved in the IRE1 or ATF6 pathways, we hypothesized that the PERK/eIF2α axis is the major UPR branch involved in maintaining rhodopsin homeostasis under ER stress. To test this hypothesis, we first generated a null allele of atf6 (atf61) using the CRISPR/CAS9 technique (Fig. S2, C and D). Resulting mutants were viable. A loss of function mutation for the ire1 gene (ire1f02170) was already available (Coelho et al., 2013). Unlike we saw for the perk mutant, levels of Rh1P37H and endogenous Rh1 were unaffected in homozygous ire1f02170 or atf61 mutants compared with Rh1P37H-GFP flies (Fig. S2, E–H).
Figure S2.
Levels of Rh1P37H and endogenous Rh1 were unaffected by mutations in the ire1 and atf6. (A) Schematic of atf4 deletion through sgRNA targeting. Organization of the atf4 locus and the expected structure of the deletion allele atf4KO are shown. Orange boxes represent the coding region. The positions of the sgRNA pair and the DNA primers used for PCR (arrows, pF and pR) are indicated. (B) Verification of the atf4KO locus by DNA sequencing. The atf4KO mutation inserts 52 bp and eliminates 462 bp within the atf4 locus. (C) Schematic of atf6 deletion through sgRNA targeting and Flp/FRT recombination. Organization of the atf6 locus and the structure of atf6FRT and atf61 is shown. Briefly, two FRT sites (red) were inserted into the atf6 locus using CRISPR/Cas9-mediated homologous recombination. The atf6FRT knock-in flies, were cross with hs-flp lines to delete the DNA fragments between the two FRT sites. PCR primers (arrows, pF and pR) were used to verify the atf61 flies. (D) Verification of the atf61 locus by DNA sequencing. The atf61 mutation eliminates 4,440 bp within the atf6 FRT locus. (E and F) Western blot of heads dissected from wild-type (ey-flp Rh1-RFP; Rh1P37H-GFP) and ire1 mutant (ey-flp Rh1-RFP; FRT82B Rh1P37H-GFP ire1f02170/FRT82B GMR-hid CL) flies against Rh1P37H-GFP and Rh1 were shown (E) and quantified (F). Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1 (one-way ANOVA, Sidak’s multiple comparisons test). (G and H) Western blot analysis of Rh1P37H-GFP and endogenous Rh1 in homozygous atf61 (atf61; Rh1P37H-GFP) null mutant heads. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01 (one-way ANOVA, Sidak’s multiple comparisons test). Source data are available for this figure: SourceData FS2.
To further demonstrate that the PERK/eIF2α pathway is the major UPR axis regulating rhodopsin homeostasis in the Rh1P37H model, we expressed eIF2αS51A to abolish PERK-dependent eIF2α phosphorylation. Consistent with data from the perk12 and eIF2Bα39 mutants, expression of eIF2αS51A led to the accumulation of Rh1P37H-GFP and reduction of endogenous Rh1 (Fig. 3, A and B). Phosphorylated eIF2α inhibits the translation of most proteins, but selectively activates translation of the transcription factor ATF4 (Fawcett et al., 1999; Harding et al., 2000a). As we have previously shown, ATF4 expression is induced in the Rh1P37H model (Fig. 1, E and F). We then tested whether induction of ATF4 is involved in the regulation of rhodopsin homeostasis. We first generated a null allele of atf4 (atf4KO) by deleting a 462 base pair fragment using CRISPR/CAS9 (Fig. S2, A and B). However, knocking out atf4 did not result in the accumulation of Rh1P37H-GFP or reduced levels of endogenous Rh1 (Fig. 3, C and D). Moreover, continuous expression of ATF4 without its translational regulation sequence under a ubiquitously expressed da (daughterless) promotor failed to rescue the accumulation of Rh1P37H-GFP and reduction of Rh1 caused by loss of perk (Fig. 3, E and F). These data demonstrated that the PERK/eIF2 pathway regulates homeostasis of rhodopsin under chronic ER stress independent of ATF4.
Figure 3.
The PERK/eIF2α signaling pathway maintains Rh1 homeostasis independent of ATF4. (A) Western blot analysis of Rh1P37H-GFP and endogenous Rh1 in eIF2αS51A (GMR-Gal4/UAS-eIF2αS51A Rh1P37H-GFP) and eIF2α (GMR-Gal4/UAS-eIF2α Rh1P37H-GFP) heads. Flies with perkRNAi expression (GMR-Gal4/UAS-perkRNAi Rh1P37H-GFP) were used as a positive control. (B) Quantification of Rh1P37H-GFP and endogenous Rh1 levels. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). (C and D) Western blot showed that levels of both Rh1P37H-GFP and endogenous Rh1 were unchanged in atf4KO (atf4KO; Rh1P37H-GFP) mutant flies, compared with wild-type control (Rh1P37H-GFP). The perk12 and eIF2Bα39 flies were used as positive controls. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). (E and F) Western blot analysis showed that continuous expression of ATF4 failed to restore Rh1 homeostasis in perk knocked down flies. Levels of Rh1P37H-GFP and wild-type Rh1 were examined (E) and quantified (F) in perk knocked down flies (GMR-Gal4/UAS-perkRNAi Rh1P37H-GFP) that expressed atf4 (da-atf4). Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). Source data are available for this figure: SourceData F3.
Over-activation of the IRE1/XBP1 axis is involved in reducing wild-type Rh1
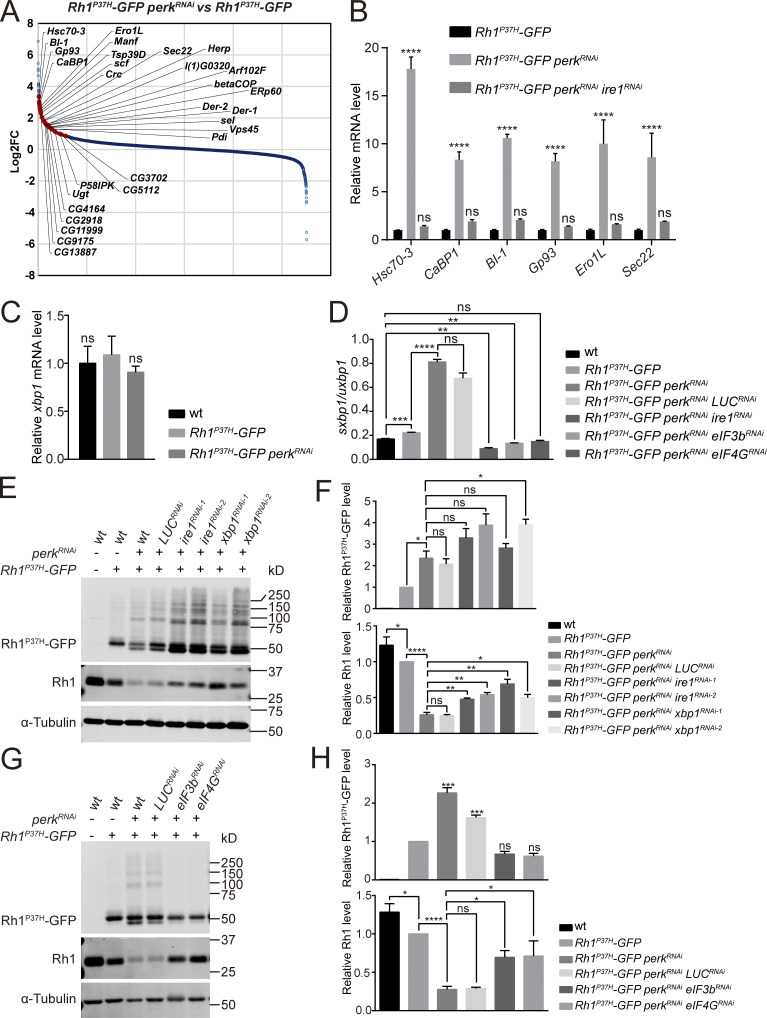
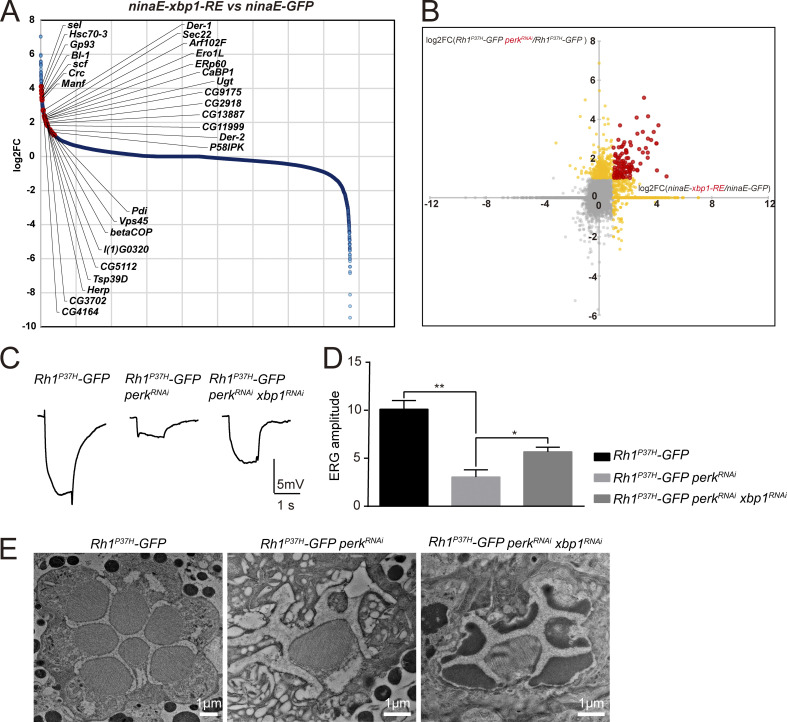
To identify factors that act downstream of PERK/eIF2α to help maintain rhodopsin homeostasis, we performed an RNA-seq analysis to identify genes that are up- or down-regulated in perk mutants. As with the perk12 mutant, knocking down perk by RNAi reduced endogenous Rh1 levels and increased levels of Rh1P37H-GFP, although to a lesser extent (Fig. 4 E). By contrast, expressing perkRNAi in a wild-type Rh1 background did not affect Rh1 homeostasis. We therefore compared the transcriptomes of Rh1P37H-GFP perkRNAi and Rh1P37H-GFP flies (Fig. 4 A). Most genes that were strongly upregulated by expression of perkRNAi were targets of the IRE1/XBP1 pathway (Hollien and Weissman, 2006). To confirm this, we used RNA-seq to identify genes up-regulated by spliced XBP1. We generated ninaE-xbp1-RE flies in which the RE form of xbp1 (the version of xbp1 that has been spliced by IRE1) is expressed in photoreceptor cells using the ninaE promotor. Importantly, most genes that were upregulated in perk mutant retinas were also induced by ninaE-xbp1-RE (Fig. S3, A and B). To further validate the RNA-seq results, we used RT-qPCR to confirm that the major IRE1/XBP1-induced genes (including Hsc70-3, CaBP1, BI-1, Gp93, Ero1L, and Sec22) were upregulated in the Rh1P37H-GFP perkRNAi retina. Importantly, upregulation of these genes was completely reversed by knocking down ire1 using ire1RNAi, further suggesting that the IRE1/XBP1 axis is activated when the PERK/eIF2α pathway is blocked during ER stress (Fig. 4 B). Since total xbp1 mRNA levels were unaffected by perkRNAi (Fig. 4 C), we next asked whether the splicing of xbp1 mRNA by IRE1 was induced upon loss of perk. We used qPCR to quantify the spliced (sxbp1) and unspliced (uxbp1) forms of xbp1. The ratio of sxbp1/uxbp1 was slightly increased in Rh1P37H-GFP flies, reflecting the mild induction of ER stress. By contrast, the sxbp1/uxbp1 ratio dramatically increased in Rh1P37H-GFP perkRNAi flies. This increase was totally abolished by ire1RNAi, further confirming that IRE1 is strongly activated when PERK is blocked during Rh1P37H-induced ER stress (Fig. 4 D).
Figure 4.
Activation of the IRE1/XBP1 axis is involved in degrading wild-type Rh1. (A) Transcriptome comparisons between retinas of Rh1P37H-GFP perkRNAi (GMR-Gal4/UAS-perkRNAi Rh1P37H-GFP) and Rh1P37H-GFP flies. The genes strongly upregulated by spliced XBP1 are indicated by red dots. (B) qPCR analysis of Hsc70-3, CaBP1, BI-1, Gp93, Ero1L, and Sec22 to confirm the RNA-seq results. Error bars indicate SEM (n = 3); ns, not significant, ****P < 0.0001 (two-way ANOVA, Sidak’s multiple comparisons test). The mRNAs were prepared from dissected retina of young (∼1-d-old) flies with indicated genotypes. (C) qPCR analysis of the total mRNA levels of xbp1 in wt, Rh1P37H-GFP, and Rh1P37H-GFP perkRNAi retina. Error bars indicate SEM (n = 3); ns, not significant (one-way ANOVA, Sidak’s multiple comparisons test). (D) The sxbp1 (spliced form)/uxbp1 (unspliced form) ratio quantified from qPCR analysis. The mRNAs were prepared from dissected retina of ∼1-d-old flies with indicated genotypes. Error bars indicate SEM (n = 3); ns, not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). (E and F) Western blot analysis of Rh1P37H-GFP and endogenous Rh1 showed that expressing ire1RNAi or xbp1RNAi significantly blocked the reduction of wild-type Rh1 by knocking down perk in Rh1P37H-GFP models. Two independent ire1RNAi and xbp1RNAi lines were used, and LUCRNAi was used as a control. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). (G and H) Western blot analysis of Rh1P37H-GFP and endogenous Rh1 showed that blocking translation by eIF3bRNAi and eIF4GRNAi suppressed both accumulation of Rh1P37H-GFP and reduction of wild-type Rh1 in Rh1P37H-GFP perkRNAi flies. LUCRNAi was used as a negative control, and α-tubulin was used as a loading control. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). Source data are available for this figure: SourceData F4.
Figure S3.
Overlap of upregulated genes in Rh1P37H-GFP perkRNAi and ninaE-xbp1-RE retinas. (A) Transcriptome profiling was used to compare gene expression between retina expressing XBP1-RE (ninaE-xbp1-RE) and GFP (ninaE-GFP) flies. Retinas were dissected from 1-d-old flies. Genes strongly upregulated by spliced XBP1 are indicated by red dots. (B) Analysis of upregulated genes in two RNA-seq experiments (Rh1P37H-GFP perkRNAi vs. Rh1P37H-GFP and ninaE-xbp1-RE vs. ninaE-GFP). Genes upregulated in both Rh1P37H-GFP perkRNAi and ninaE-xbp1-RE flies are indicated by red dots. (C and D) ERG recordings show that expressing xbp1RNAi restored visual responses in Rh1P37H-GFP perkRNAi flies. 5-d-old flies of indicated genotypes were exposed to a 1-s pulse of orange light after 2 min of dark adaptation. At least six flies were used for statistical analyses. Error bars indicate SEM (n = 6); *P < 0.1, **P < 0.01 (one-way ANOVA, Sidak’s multiple comparisons test). (E) Representative TEM images of tangential sections through the eye of Rh1P37H-GFP perkRNAi flies that express xbp1RNAi or not. Scale bar, 1 μm. Sectioned eyes were from 5-d-old flies.
PERK/eIF2α signaling generally inhibits translation, but it preferentially induces the translation of ATF4 during the UPR response. Given that the PERK/eIF2α pathway regulates rhodopsin homeostasis independent of ATF4, we speculated that PERK may negatively regulate IRE1 activity by inhibiting translation. To test this hypothesis, we re-inhibited translation in Rh1P37H-GFP perkRNAi flies by knocking down the eukaryotic translation initiation factors eIF3b and eIF4G. We found that the sxbp1/uxbp1 ratio decreased to wild-type levels (Fig. 4 D).
Since IRE1 is strongly activated in perk mutants upon ER stress, we next examined whether inhibition of the IRE1/XBP1 pathway contributes to rhodopsin homeostasis. We first knocked down ire1 or xbp1 in the background of Rh1P37H-GFP perkRNAi and found that the levels of endogenous Rh1 were increased compared with control. However, the increase of Rh1P37H-GFP in perkRNAi mutants was not alleviated, and even slightly aggravated by knocking down ire1 or xbp1 (Fig. 4, E and F). These data indicate that accumulation of mutant Rh1 and reduction of wild-type proteins are independent events regulated by different signaling pathways; the latter is mediated by IRE1/XBP1. Supporting this point, as seen when ire1 or xbp1 were knocked down, expressing eIF3bRNAi or eIF4GRNAi in the background of Rh1P37H-GFP perkRNAi restored levels of wild-type Rh1 (Fig. 4, G and H). In contrast to ire1 and xbp1 knock down, when knocking down eIF3b or eIF4G, levels of Rh1P37H-GFP in Rh1P37H-GFP perkRNAi flies was also reduced to wild-type levels, supporting the hypothesis that reductions in translation efficiency by PERK activation is the major event involved in rhodopsin homeostasis (Fig. 4, G and H). As knocking down ire1 or xbp1 specifically prevented the loss of wild-type rhodopsin, we were able to examine if the loss of wild-type rhodopsin contributed to the retinal degeneration seen in Rh1P37H-GFP perkRNAi flies. 5-d-old Rh1P37H-GFP perkRNAi flies exhibited phenotypes associated with severe retinal degeneration, including reduced ERG responses and loss of rhabdomeres (Fig. S3, C–E). Expression of xbp1RNAi ameliorated ERG responses and suppressed the loss of photoreceptor cells in Rh1P37H-GFP perkRNAi flies (Fig. S3, C–E). Considering that knocking down ire1 or xbp1 in the background of Rh1P37H-GFP perkRNAi only alleviated reduction of wild-type Rh1 without affecting Rh1P37H-GFP, loss of wild-type rhodopsin may be involved in the pathogenesis of this dominant rhodopsin disorder.
The selective autophagy of ER is involved in degradation of wild-type Rh1
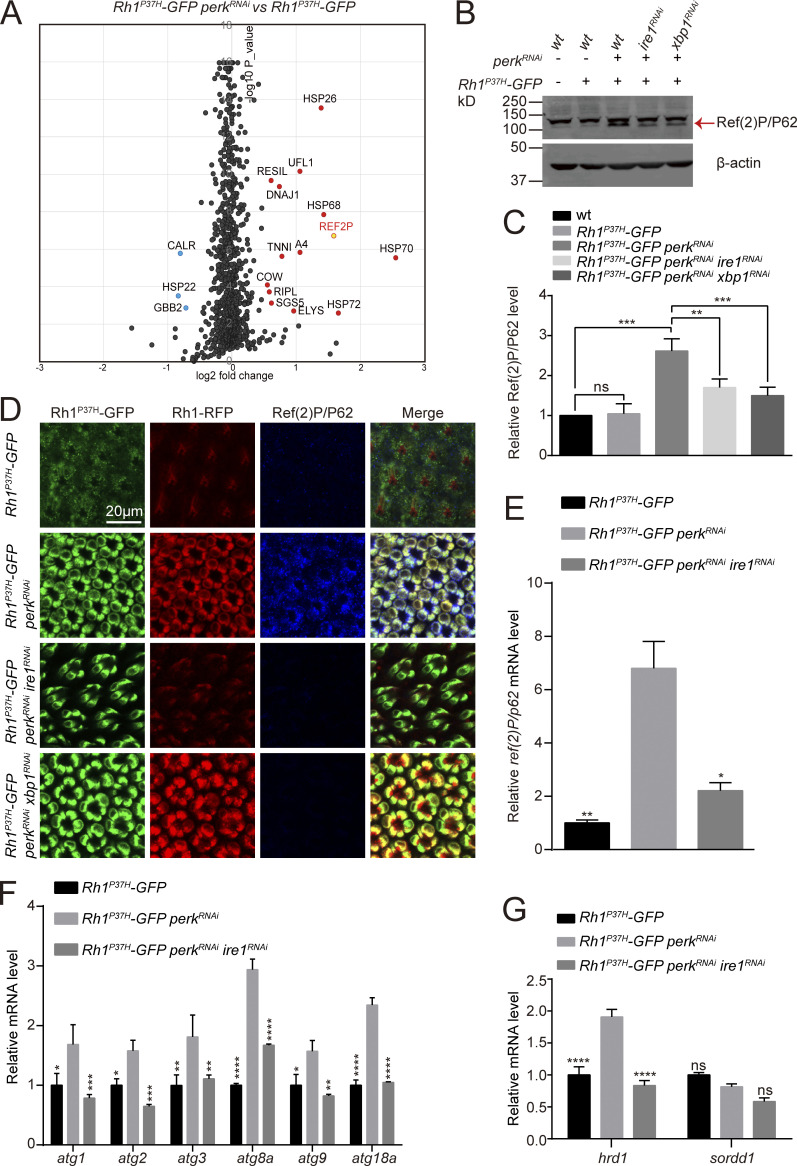
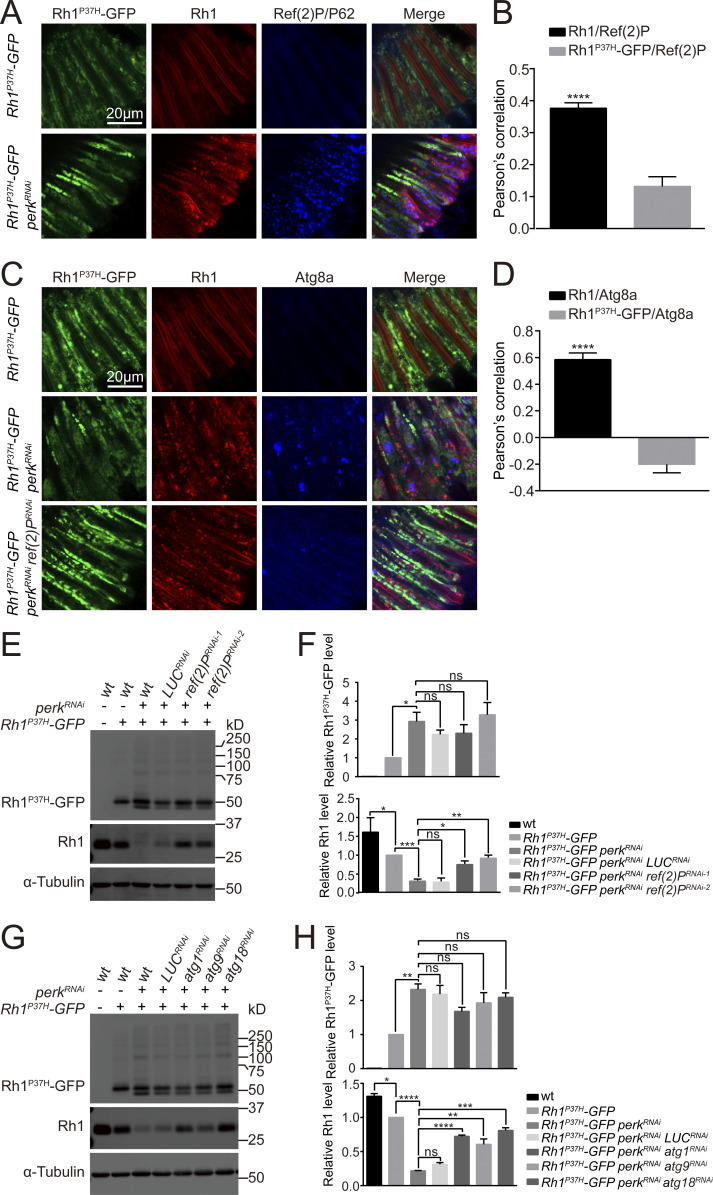
Downregulation of wild-type Rh1 by the IRE1/XBP1 signaling pathway leads to the loss of rhodopsin homeostasis in Rh1P37H expressing cells. To investigate the mechanisms of this process, we used Tandem Mass Tag (TMT)–LC MS/MS to compare individual protein levels between Rh1P37H-GFP perkRNAi and Rh1P37H-GFP retinas. We identified 14 proteins that were greatly upregulated (P value <0.01) when perk was knocked down (Fig. 5 A). We then screened these 14 candidate genes by knocking them down in photoreceptor cells of Rh1P37H-GFP perkRNAi flies. Only one gene, ref(2)P/p62, blocked the reduction of Rh1 when knocked down (Fig. 6, E and F). The ref(2)P/p62 gene encodes an evolutionary conserved autophagy adaptor in Drosophila, indicating that autophagy is modulated in this context. We first verified that the Ref(2)P/P62 protein was upregulated in Rh1P37H-GFP perkRNAi mutant retina by Western blotting and immunostaining. Importantly, this increase in Ref(2)P/P62 proteins was abolished by expressing ire1RNAi or xbp1RNAi (Fig. 5, B–D). Moreover, ref(2)P/p62 mRNA levels were also elevated in Rh1P37H-GFP perkRNAi flies; this could also be reversed by ire1RNAi (Fig. 5 E). These data indicated that the autophagy pathway may be induced by blocking the PERK pathway in Rh1P37H expressing photoreceptor cells. Consistent with this hypothesis, mRNA levels of several autophagy genes including atg1, atg2, atg3, atg8a, atg9, and atg18a, were also increased by perkRNAi in the context of Rh1P37H-induced ER stress. Knocking down ire1 suppressed these inductions (Fig. 5 F). We further measured levels of hrd1 and sordd1 mRNA, which encode two ER-associated ubiquitin ligases, and found that IRE1/XBP1 signaling also induced the expression of hrd1 but not sordd1 (Fig. 5 G).
Figure 5.
Autophagy is induced by blocking PERK in Rh1P37H photoreceptor cells. (A) Proteomic profiling comparing protein levels in retinas of Rh1P37H-GFP perkRNAi (GMR-Gal4/UAS-perkRNAi Rh1P37H-GFP) and Rh1P37H-GFP flies (Tandem Mass Tag-LC MS/MS assay). A total of 1,339 proteins were confidently identified (at least two unique peptides per protein). A subset of proteins up- or downregulated in Rh1P37H-GFP perkRNAi cells are highlighted by protein identification. (B and C) Western blotting confirmed that Ref(2)P/P62 was upregulated in Rh1P37H-GFP perkRNAi flies. This was abolished by expression of ire1RNAi and xbp1RNAi. Ref(2)P/P62 protein is indicated by the red arrow. Error bars indicate SEM (n = 6); ns, not significant, **P < 0.01, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). (D) Tangential views of retina expressing Rh1P37H-GFP (green) and Rh1-RFP (red) with perkRNAi and/or ire1RNAi/xbp1RNAi staining against Ref(2)P/P62 (blue). Scale bar, 20 μm. (E) qPCR analysis of ref(2)P/p62 mRNA levels in Rh1P37H-GFP perkRNAi and Rh1P37H-GFP perkRNAi ire1RNAi retinas compared with Rh1P37H-GFP controls. Error bars indicate SEM (n = 3); *P < 0.1, **P < 0.01 (one-way ANOVA, Sidak’s multiple comparisons test). (F) qPCR analysis showed that mRNA levels of autophagy-related genes (including atg1, atg2, atg3, atg8a, atg9, and atg18a) were upregulated in the retina of Rh1P37H-GFP perkRNAi flies, compared with Rh1P37H-GFP and Rh1P37H-GFP perkRNAi ire1RNAi retina. Error bars indicate SEM (n = 3); *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001 (two -way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. (G) qPCR analysis showed that mRNA levels of the ER-associated E3 ligase, hrd1 (bot not sordd1) was upregulated in the retina of Rh1P37H-GFP perkRNAi flies, compared with Rh1P37H-GFP and Rh1P37H-GFP perkRNAi ire1RNAi retina. Error bars indicate SEM (n = 3); ns, not significant, ****P < 0.0001 (two-way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. Source data are available for this figure: SourceData F5.
Figure 6.
Wild-type Rh1 but not Rh1P37H is degraded through autophagy. (A–D) Wild-type Rh1, but not Rh1P37H colocalized with Ref(2)P/P62 and Atg8a in Rh1P37H-GFP perkRNAi flies. (A) Longitudinal views of photoreceptor cells labeled for GFP (Rh1P37H-GFP, green), RFP (Rh1-RFP, red), and Ref(2)P/P62 (blue). Scale bar, 20 μm. (B) Quantification of the co-localization between Rh1P37H-GFP or Rh1-RFP and Ref(2)P/P62 in Rh1P37H-GFP perkRNAi photoreceptor cells. Error bars indicate SEM (n = 3); ****P < 0.0001 (Student’s unpaired t test). (C) Immune staining of photoreceptor cells against GFP (Rh1P37H-GFP, green), RFP (Rh1-RFP, red), and Atg8a (blue). Scale bar, 20 μm. (D) Quantification of the co-localization between Rh1P37H-GFP or Rh1-RFP and Atg8a in Rh1P37H-GFP perkRNAi photoreceptor cells. Error bars indicate SEM (n = 3); ****P < 0.0001 (Student’s unpaired t test). (E and F) Western blotting showed that knocking down ref(2)P/p62 significantly blocked the reduction of wild-type Rh1 without affecting the accumulation of Rh1P37H-GFP in Rh1P37H-GFP perkRNAi retina. Two independent ref(2)P/p62RNAi lines were used, and LUCRNAi was used as a control. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). (G and H) Western blot analysis against Rh1P37H-GFP and endogenous Rh1 showed that atg1RNAi, atg9RNAi, and atg18RNAi suppressed perkRNAi-mediated decreases in wild-type Rh1 in Rh1P37H-GFP photoreceptor cells. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). LUCRNAi was used as a control line, and α-tubulin was used as a loading control. 1-d-old flies of indicated genotypes were used. Source data are available for this figure: SourceData F6.
To further confirm that autophagy is induced by loss of perk in Rh1P37H-expressing photoreceptor cells, we labeled retinas for the autophagy marker, Atg8a, and Ref(2)P/P62. Both proteins accumulated in photoreceptor cells expressing both Rh1P37H-GFP and perkRNAi, but not in cells expressing Rh1P37H-GFP alone (Fig. 6, A and C). Since autophagy is induced by the mutation of perk through IRE1 in Rh1P37H expressing photoreceptors, it is possible that wild-type Rh1, but not mutant Rh1P37H, is degraded by autophagy. Supporting this, endogenous Rh1 was detected in cytosolic puncta that colocalized with Ref(2)P/P62 and Atg8a in Rh1P37H-GFP perkRNAi flies. By contrast, Rh1P37H-GFP was not detected in Ref(2)P/P62- and Atg8a-positive puncta (Fig. 6, A–D). In TEM images we also observed the presence of autophagosome structures in Rh1P37H-GFP perk12 photoreceptor cells, but not in Rh1P37H-GFP photoreceptor cells (Fig. S4 A). Knocking down ref(2)P/p62 and autophagy-associated genes including atg1, atg9, and atg18 largely increase wild-type Rh1 levels in Rh1P37H-GFP perkRNAi flies. As was seen when we blocked the IRE1/XBP1 signaling pathway, disrupting autophagy did not affect Rh1P37H-GFP levels (Fig. 6, E–H). Moreover, knocking down ref(2)P/p62 reduced the number of Atg8a-positive puncta without affecting the aggregation of wild-type Rh1. This suggests that this selective autophagy occurs downstream of rhodopsin misfolding and/or ubiquitination (Fig. 6 C).
Figure S4.
ER proteins were degraded through autophagy in Rh1P37H-GFP perkRNAi photoreceptor cells. (A) TEM images of tangential sections of compound eyes from 5-d-old wt (w1118), Rh1P37H-GFP, and Rh1P37H-GFP perkRNAi flies. Autophagosome are indicated by red arrows. Scale bars, 2 μm and 500 nm. All flies were in white eye background and raised under 12 h light/12 h dark cycles. (B–H) Western blotting against GFP to examine the effects of perkRNAi on proteins with different subcellular localizations. ER-GFP was decreased in Rh1P37H-GFP perkRNAi flies, whereas Rh1P37H-GFP perkRNAi only slightly reduced mito-GFP and cytosolic GFP. Error bars indicate SEM (n = 6); ns, not significant, *P < 0.1, **P < 0.01, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. (I) qPCR analysis showed that mRNAs encoding fly homologs of mammalian ER-phagy receptors (trp1/sec62 and atl/atl3), but not homologs of mito-phagy receptors (nipsnap and phb2), were upregulated in the retina of Rh1P37H-GFP perkRNAi flies, compared with Rh1P37H-GFP and Rh1P37H-GFP perkRNAi ire1RNAi retina. Error bars indicate SEM (n = 3); ns, not significant, ****P < 0.0001 (two-way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. (J) Accumulation of ubiquitinated proteins in membrane extracts of Rh1P37H-GFP perkRNAi flies. The membrane fraction was purified via centrifuge and labeled for ubiquitin and calnexin. Source data are available for this figure: SourceData FS4.
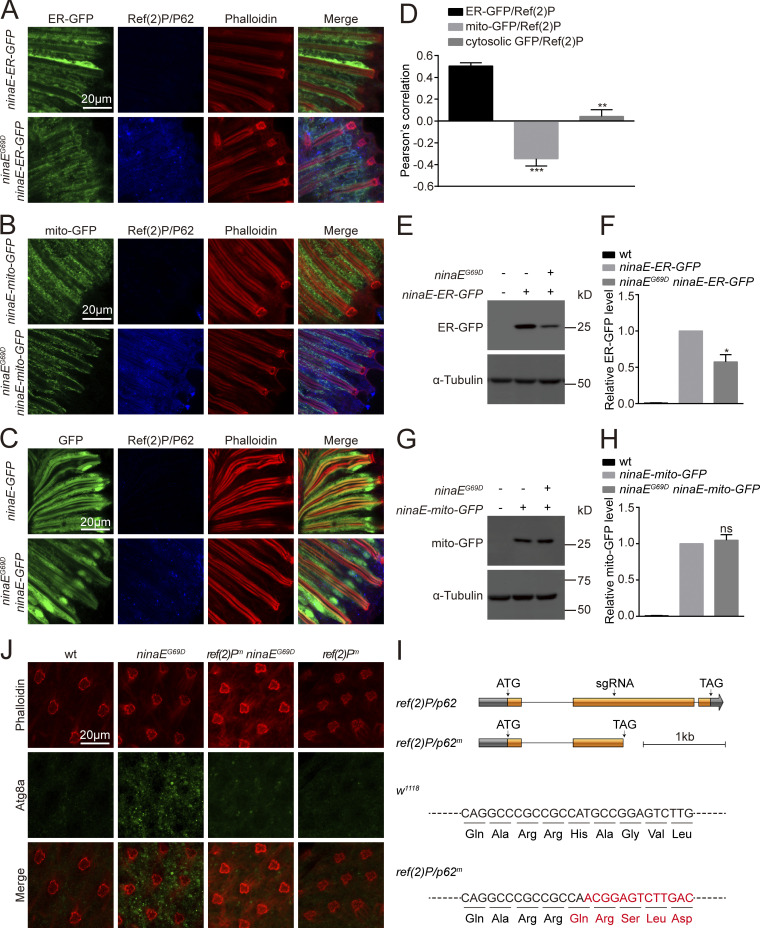
Since autophagy is a general degradation system involved in the turnover of proteins in multiple cellular components, we asked if the autophagy observed in Rh1P37H-GFP perk12 photoreceptor cells is selective for components of the ER. We drove expression of GFP reporters specific for different subcellular compartment (ER, mitochondria, and cytosol) via the ninaE promoter in Rh1P37H-GFP perkRNAi flies. Levels of ER-GFP were dramatically reduced in Rh1P37H-GFP perkRNAi flies, whereas reporters specific for mitochondria (mito-GFP) or cytosolic GFP were only slightly reduced. This may reflect the unhealthy state of photoreceptor cells under chronic ER stress (Fig. S4, B–H). In addition to Ref(2)P/P62, two homologs of ER-phagy receptors functioning in mammalian cells, trp1/sec62 (Fumagalli et al., 2016) and atl/atl3 (Chen et al., 2019), were also induced in Rh1P37H-GFP perkRNAi flies. This also could be reversed by knocking down ire1 (Fig. S4 I). In contrast, the expression of two fly homologs of mammalian mitophagy receptors, nipsnap (Princely Abudu et al., 2019) and phb2 (Wei et al., 2017), were not affected (Fig. S4 I). These data suggest that selective autophagy of ER is induced when the PERK pathway is blocked, and that this autophagy is responsible for reducing wild-type Rh1 in Rh1P37H-GFP perkRNAi flies.
Rh1P37H-GFP is degraded by the UPS system, which is impaired in perk mutant cells
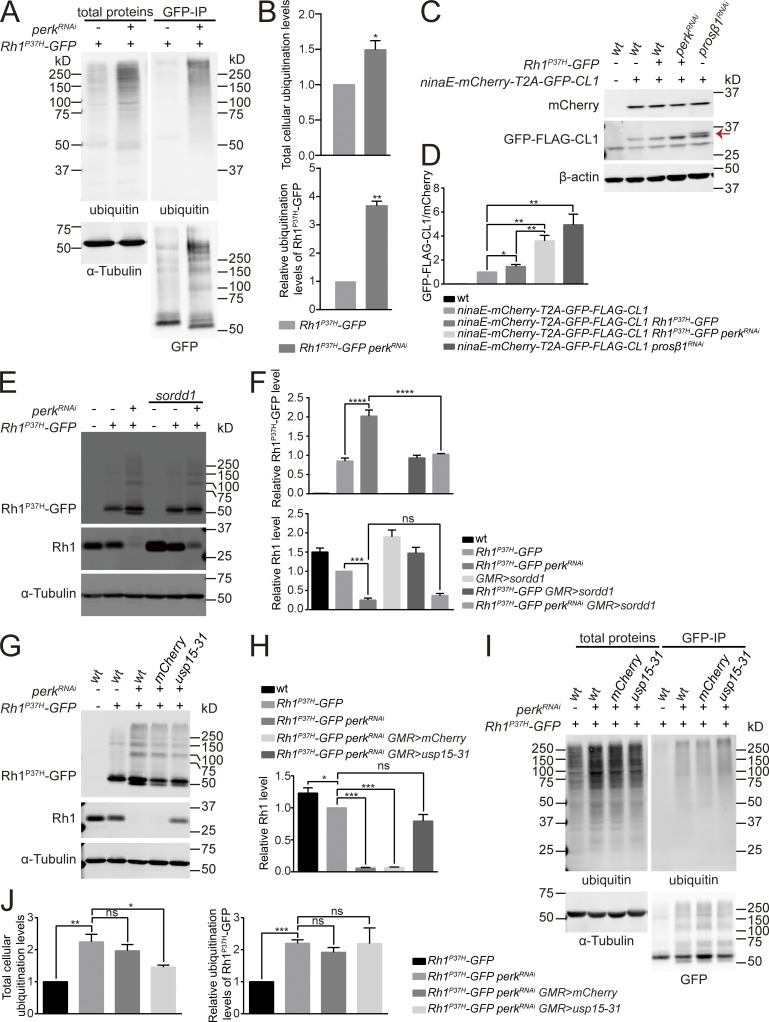
Misfolded membrane proteins are recognized by ER chaperons and removed from the ER via a ubiquitin-proteasome system (UPS)-mediated degradation process called ER-associated degradation (ERAD; Meusser et al., 2005; Vembar and Brodsky, 2008). Therefore, the accumulation of mutant Rh1P37H protein may result from an impairment of the ubiquitin/proteasome degradation system. We first tested if the ubiquitination machinery was disrupted in Rh1P37H-GFP perkRNAi photoreceptor cells. However, both total cellular ubiquitinated proteins and ubiquitinated Rh1P37H-GFP accumulated in Rh1P37H-GFP perkRNAi flies compared with Rh1P37H-GFP controls (Fig. 7, A and B). These data indicate that proteasome activity in Rh1P37H-GFP perkRNAi photoreceptor cells may be reduced. To test this hypothesis, we used a proteasome activity reporter, GFP-Flag-Cl1, which contains a short degron fragment (CL1) that is degraded by the ubiquitin-proteasome system (Gilon et al., 1998; Nonaka and Hasegawa, 2009). GFP-Flag-Cl1 was linked to mCherry (as an internal control) via a self-cleaving peptide T2A. The reporter was then expressed in photoreceptor cells via the ninaE promoter. Compared with wild-type flies, GFP-Flag-CL1 levels were slightly increased in Rh1P37H-GFP flies. Levels of GFP-Flag-CL1 were dramatically elevated when the proteasomal subunit prosβ1 was knocked down (Fig. 7, C and D). Importantly, as seen with blocking the proteasomal subunit, perk mutation largely stabilized GFP-FLAG-CL1 in Rh1P37H-GFP flies (Fig. 7, C and D). These data suggest that the UPS system is impaired when PERK is blocked during ER stress. Moreover, expression of the E3 ligase SORDD1 (Xu et al., 2020) in Rh1P37H-GFP perkRNAi photoreceptor cells prevented the accumulated Rh1P37H-GFP (Fig. 7, E and F) but did not affect wild-type Rh1 levels. This is consistent with SORDD1 only being involved in the degradation of misfolded rhodopsin (Xu et al., 2020).
Figure 7.
The ubiquitin-proteasome system (UPS) is impaired upon blocking PERK in adRP photoreceptors. (A) Accumulation of ubiquitinated proteins in Rh1P37H-GFP perkRNAi flies. Head lysate of Rh1P37H-GFP and Rh1P37H-GFP perkRNAi flies were immunoprecipitated with anti-GFP beads and stained against ubiquitin and GFP. (B) Quantification of total cellular ubiquitination levels (upper panel), and relative ubiquitination levels of Rh1P37H-GFP (lower panel) in A. Error bars indicate SEM (n = 3); *P < 0.1, **P < 0.01 (Student’s unpaired t test). (C) Western blotting against GFP and mCherry to examine proteasome activity in Rh1P37H-GFP perkRNAi flies. The ninaE promoter was used to drive mCherry-T2A-GFP-FLAG-Cl1 expression. Knock down of the proteasome subunit by prosβ1RNAi was used as a positive control. GFP-FLAG-CL1 protein is indicated by the red arrow. (D) Quantification of GFP-FLAG-CL1 and mCherry ratio in C. Error bars indicate SEM (n = 3); *P < 0.1, **P < 0.01 (one-way ANOVA, Sidak’s multiple comparisons test). (E and F) Western blot analysis showed that overexpression of SORDD1 in photoreceptor cells reduced Rh1P37H-GFP specifically upon loss of perk without affecting the Rh1 levels. Error bars indicate SEM (n = 3); ns, not significant, ***P < 0.001, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). (G and H) Western blot analysis showed that USP15-31 restored Rh1 levels in Rh1P37H-GFP perkRNAi flies without affecting the levels of Rh1P37H-GFP. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). (I) Overexpression of USP15-31 reduced the accumulation of ubiquitinated proteins in Rh1P37H-GFP perkRNAi flies, without affecting the ubiquitination levels of Rh1P37H-GFP. Head lysates of Rh1P37H-GFP, Rh1P37H-GFP perkRNAi, Rh1P37H-GFP perkRNAi mCherry (GMR-Gal4/UAS-mCherry;UAS-perkRNAi/Rh1P37H-GFP) and Rh1P37H-GFP perkRNAi usp15-31 (GMR-Gal4/UAS-usp15-31;UAS-perkRNAi/Rh1P37H-GFP) flies were immunoprecipitated with anti-GFP beads and stained against ubiquitin and GFP. (J) Quantification of total cellular ubiquitinated proteins (left panel), and relative ubiquitination levels of Rh1P37H-GFP (right panel) in I. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1, **P < 0.01, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. Source data are available for this figure: SourceData F7.
The Ref(2)P/P62 protein is an adaptor that binds ubiquitinated proteins and autophagy components, thereby serving as a linker between the autophagy machinery and its targets. Considering the fact that total ubiquitination levels increased in Rh1P37H-GFP perkRNAi flies, we reasoned that the accumulation of ubiquitinated ER proteins initiated the ER-phagic degradation of wild-type Rh1. Consistent with induction of the ERAD ubiquitin ligase, Hrd1, ubiquitinated membrane proteins accumulated in Rh1P37H-GFP perkRNAi flies. This could be reversed by knocking down ire1 (Fig. S4 J). To further test this hypothesis, we overexpressed the general cytosolic deubiquitinase, USP15-31, in Rh1P37H-GFP perkRNAi flies and found that USP15-31 restored levels of wild-type Rh1 in Rh1P37H-GFP perkRNAi flies without affecting the levels of Rh1P37H-GFP (Fig. 7, G and H). Further, total cellular ubiquitination levels were largely decreased in Rh1P37H-GFP perkRNAi flies when USP15-31 was overexpressed, whereas ubiquitination levels of Rh1P37H-GFP were unaffected (Fig. 7, I and J). These data provided further evidence that Rh1 degradation is regulated by Ref(2)P/P62-mediated selective autophagy, which is different from UPS-mediated degradation of Rh1P37H-GFP.
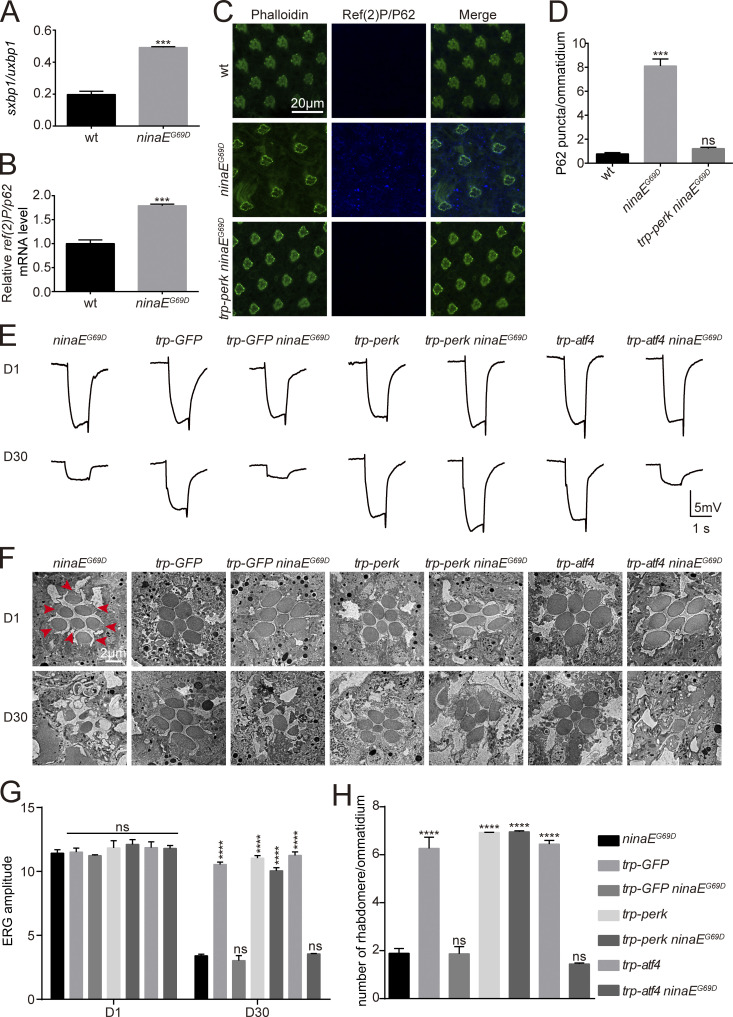
PERK prevents retinal degeneration in the ninaEG69D model of adRP
Since inhibiting PERK induced ER-phagic degradation of wild-type Rh1, and increased the cytotoxicity of misfolded Rh1P37H, it is possible that induction of PERK could suppress autophagy and alleviate retinal degeneration associated with mutations in rhodopsin. To test this hypothesis, we used a classic adRP model in which the ninaEG69D mutation leads to age-dependent degeneration of photoreceptor cells (Colley et al., 1995; Kurada and O'Tousa, 1995). We first examined if autophagy is induced by the ninaEG69D mutation. Consistent with previous results using Rh1P37H, the sxbp1/uxbp1 ratio was increased in ninaEG69D flies, indicating activation of the UPR (Fig. 8 and Fig. 9 A). ninaEG69D photoreceptor cells also exhibited large increases in ref(2)P/p62 mRNA levels, as well as Ref(2)P/P62- and Atg8-positive puncta, compared with wild-type controls (Fig. 9, B–D and Fig. S5 J). Moreover, ER-GFP but not mito-GFP or cytosolic GFP colocalized with Ref(2)P/P62 in ninaEG69D flies (Fig. S5, A–D), and levels of ER-GFP but not mito-GFP protein were significantly reduced in ninaEG69D flies (Fig. S5, E–H). To test the role of Ref(2)P/P62 in inducing autophagy in ninaEG69D photoreceptor cells, we used the CRISPR-CAS9 system to generate a ref(2)P/p62 mutant fly (Fig. S5 I). The ref(2)Pm mutation itself did not affect autophagy in photoreceptor cells. However, loss of Ref(2)P/P62 in ninaEG69D photoreceptor cells largely abolished the formation of Atg8a puncta (Fig. S5 J). These data indicate that the selective autophagy of ER is induced in the ninaEG69D model of adRP and is involved in general degradation of the ER compartment. Importantly, overexpressing PERK in photoreceptor cells under control of the endogenous trp (transient receptor potential) promoter greatly reduced levels of Ref(2)P/P62 in aged ninaEG69D photoreceptor cells, compared to wild-type controls (Fig. 9, C and D). ninaEG69D flies exhibited phenotypes consisted with severe retinal degeneration including reduced ERG responses, and loss of rhabdomeres and photoreceptor cells ∼30 d after eclosion (Fig. 9, E–H). Expression of PERK in photoreceptor cells completely restored ERG responses and prevented the loss of photoreceptor cells in 30-d-old ninaEG69D flies (Fig. 9, E–H). In contrast, overexpression of ATF4 did not affect the loss of ERG and photoreceptor cells in the ninaEG69D mutants (Fig. 9, E–H). Consistent with our previous results, these data demonstrate that PERK suppresses adRP independent of ATF4.
Figure 8.
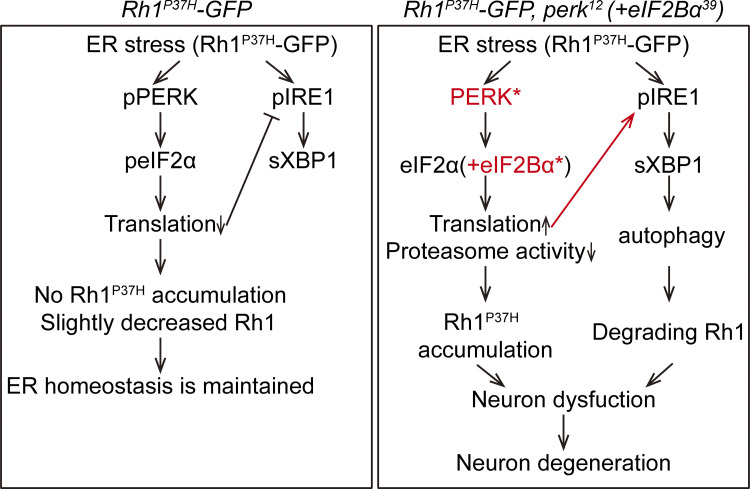
Working model of cell protective role of PERK in prolonged ER stress triggered by misfolded Rh1P37H. On the left we illustrate that PERK signaling plays a central role in maintaining rhodopsin homeostasis and cellular function in cells expressing Rh1P37H. On the right we show that deficiency in the PERK pathway (perk12 or eIF2Bα39 mutation) de-inhibits translation and over-activates IRE1, leading to the accumulation of Rh1P37H due to insufficient proteasome activity and degradation of wild-type Rh1 through induction of autophagy. This ultimately causes neuron dysfunction and degeneration.
Figure 9.
PERK prevents retinal degeneration in the ninaEG69D model of adRP. (A and B) qPCR analysis of the sxbp1/uxbp1 ratio (A) and mRNA levels of ref(2)P/p62 (B) demonstrated that the IRE1/XBP1/autophagy axis was activated in ninaEG69D photoreceptor cells. Error bars indicate SEM (n = 3); ***P < 0.001 (Student’s unpaired t test). (C and D) Immunostaining of photoreceptor cells against Ref(2)P/P62 showed an increase of Ref(2)P/P62 puncta in ninaEG69D photoreceptor cells. This was abolished by ectopic expression of PERK. Phalloidin served as an ommatidia maker. Error bars indicate SEM (n = 3); ns, not significant, ***P < 0.001 (one-way ANOVA, Sidak’s multiple comparisons test). (E) Representative ERG recordings of 1-d-old and 30-d-old flies showed that PERK but not ATF4 prevented the loss of visual response in ninaEG69D flies. Flies with indicated genotypes were exposed to a 1-s pulse of orange light after 2 min of dark adaptation. (F) Representative TEM images of 1-d-old and 30-d-old eye tangential sections. Genotypes are indicated. Rhabdomeres are indicated by red arrows. Scale bar, 2 μm. (G) Statistical analysis of the amplitude of ERG recordings for 1-d-old and 30-d-old flies from E. Error bars indicate SEM (n = 6); ns, not significant, ****P < 0.0001 (two-way ANOVA, Sidak’s multiple comparisons test). (H) Quantification of the number of rhabdomeres per ommatidia in F. Sections from three 30-d-old flies of each genotype were used for quantification. ns, not significant, ****P < 0.0001 (one-way ANOVA, Sidak’s multiple comparisons test). All flies were in white eye background, and were raised in 12-h light/12-h dark (L/D) cycles at 25°C.
Figure S5.
ER-phagy was induced in the ninaEG69D model of adRP. (A–D) ER-GFP, but not mito-GFP or cytosolic GFP colocalized with Ref(2)P/P62 in ninaEG69D photoreceptor cells. Longitudinal views of retinas from ninaE-ER-GFP/ninaEG69D (A), ninaE-mito-GFP/ninaEG69D (B), and ninaE-GFP/ninaEG69D(C) flies labeled against GFP (green) and Ref(2)P/P62 (blue). Phalloidin (red) was used as a marker for rhabdomere. Scale bar, 20 μm. (D) Quantification of the co-localization between Ref(2)P/P62 and ER-GFP, mito-GFP, or cytosolic GFP in ninaEG69D photoreceptor cells. Error bars indicate SEM (n = 3); **P < 0.01, ***P < 0.001 (Student’s unpaired t test). (E–H) Western blotting against GFP to examine the levels of ER-GFP (E and F) and mito-GFP (G and H) in the ninaEG69D background. Error bars indicate SEM (n = 3); ns, not significant, *P < 0.1 (one-way ANOVA, Sidak’s multiple comparisons test). 1-d-old flies of indicated genotypes were used. (I) Generation of ref(2)Pm flies. Organization of the ref(2)P/p62 locus is shown. A single sgRNA primer was used to generate the mutations. The ref(2)Pm frame-shift mutation was identified via DNA sequencing. (J) Immunostaining photoreceptor cells for Atg8a showed an increase in Atg8a puncta in ninaEG69D photoreceptor cells. This was abolished when the ref(2)Pm mutation was introduced. Phalloidin served as a marker for ommatidia. Source data are available for this figure: SourceData FS5.
Discussion
In conclusion, we found that PERK is important for maintaining rhodopsin homeostasis during prolonged ER stress. In photoreceptor cells expressing misfolded Rh1P37H, the continuous activation of PERK attenuates IRE1/XBP1 signaling, which is adapted to this chronic ER stress, in an eIF2α-dependent manner. However, when PERK is blocked, global translation is no longer inhibited by phosphorylated eIF2α. This increases the burden on the proteasome and reduces degradation of Rh1P37H by ERAD. By contrast, in the absence of the PERK pathway, IRE1/XBP1 signaling remains activated, inducing multiple autophagy-related genes including ref(2)P/p62. Together with the accumulation of ubiquitinated proteins due to proteasome overload, the upregulation of these autophagy-related genes induces the selective autophagy of ER. This promotes the degradation of wild-type rhodopsin that has accumulated in the ER, ultimately triggering photoreceptor cell death (Fig. 8).
The three branches of the UPR (IRE1, PERK, and ATF6) are simultaneously induced in response to cellular stress. However, in the context of chronic ER stress, the three UPR pathways are differentially affected. This is particularly true for the IRE1 and PERK branches, which may be due to interactions between these two signaling pathways. During development of plasma cells, IRE1 is robustly activated, whereas activation of PERK is suppressed by Ufbp1 (Ufm1-binding protein; Zhu et al., 2019). Under prolonged ER stress, the activities of IRE1 and ATF6 are attenuated, whereas PERK signaling persists (Lin et al., 2007). In cultured cells, translational attenuation by PERK facilitates IRE1 during the early adaptive phase of the UPR (Moore and Hollien, 2015), but during the prolonged UPR stage, IRE1 activity is attenuated by PERK/eIF2α-dependent and ATF4-independent translational inhibition to turn off IRE1 activity (Chang et al., 2018). In chronic ER stress induced by Rh1P37H, blocking PERK signaling activates xbp1 splicing and thus the transcription function of XBP1. Consistent with results seen with pharmacologically induced ER stress, this IRE1/XBP1 re-activation is mediated by loss of translational inhibition in perk mutants, as slowing translation by expressing short-hairpin RNAs against eIF3b and eIF4G suppressed XBP1 over-activation by loss of perk. This suggests the mechanisms of prolonged ER-stress adaptation through attenuation of IRE1 by PERK are conserved across species. However, in contrast to cultured cells, where pharmacological ER stress attenuates IRE1 through PERK to trigger apoptosis, in photoreceptor cells in vivo ER-stress adaptation through attenuation of IRE1 is cytoprotective. In Rh1P37H-expressing photoreceptor cells, severe cell death was induced by mutations of both perk and eIF2Bα, whereas the cells survived upon Rh1P37H expression alone. Further, IRE1-mediated xbp1 splicing was boosted, and wild-type rhodopsin was largely degraded by blocking of PERK in Rh1P37H-expressing cells. Knocking down ire1 or xbp1 completely rescued this rhodopsin loss. Therefore, our present study suggests that the downregulation of IRE1 by PERK is cytoprotective during chronic ER stress.
In both yeast and neuroblastoma cells, induction of ER stress activates autophagy in an IRE1-dependent manner (Ogata et al., 2006; Yorimitsu et al., 2006). In Drosophila neurons, ectopic overexpression of IRE1 was sufficient to induce autophagy and triggered neuron death (Yan et al., 2019). As a cellular quality control mechanism for clearance of proteins and organelles, loss or over-activated of autophagy can trigger neuronal cell death under certain pathological conditions (Hara et al., 2006; Komatsu et al., 2006; Wang et al., 2009). Besides clearing misfolded proteins, autophagy can degrade normal folded proteins or undamaged organelles derived from imbalanced proteostasis; this ultimately disrupts cellular function (Berry and Baehrecke, 2007; Doherty and Baehrecke, 2018). A previous study found that autophagy flux is elevated in RhoP23H/+ mice, and inhibiting autophagy reduces retinal degeneration caused by protein misfolding (Yao et al., 2018). However, how autophagy is induced in this adRP model remains unclear, and the molecular mechanisms linking autophagy and RhoP23H-induce photoreceptor cell degeneration remains a mystery. In our study, we first observed in the retinas of Rh1P37H-GFP perkRNAi: (1) dramatic accumulations of the autophagic substrate Ref (2)P/P62 and the autophagy marker LC3/ATG8, (2) transcriptional activation of multiple autophagy-related genes, and (3) a large increase in autophagy flux. Further evidence showed that downregulation of IRE1/XBP1 signaling reversed the induction of autophagy in Rh1P37H-GFP perkRNAi photoreceptor cells. Finally, in the fly adRP model ninaEG69D, accumulation of Ref(2)P/P62- and Atg8-positive puncta was associated with xbp1 splicing. Studies in both Drosophila and mammalian cells have demonstrated that induction of PERK/eIF2α induces autophagy (B'Chir et al., 2013; Nagy et al., 2013), indicating that PERK/eIF2α signaling could regulate autophagy through different signaling pathways. We conclude that the uncontrolled activation of IRE1/XBP1 signaling could induce autophagy under prolonged ER stress.
Degradation of misfolded ER proteins via ERAD, which involves the ubiquitin–proteasome system, is a major mechanism for maintaining proper proteostasis of the ER (Walter and Ron, 2011). Maintaining proteostasis of ER proteins may also require the second degradative system of the cell, the autophagy-lysosome pathway through selective ER autophagy, ER-phagy (Khaminets et al., 2015). Since ER-phagy was originally described, it has been shown that several ER-phagy receptors interact with the autophagy-related protein LC3/ATG8 to recruit the autophagosome and promote the selective clearance of excessive membrane portions of the ER (An et al., 2019; Fumagalli et al., 2016; Grumati et al., 2017; Khaminets et al., 2015). However, discovery of these ER-phagy receptors did not reveal whether ER-phagy is directly involved in the clearance of misfolded proteins (in parallel or complementary to ERAD), thereby playing a role in the UPR. In our present study, we present evidence that ER-phagy is induced by prolonged ER stress through imbalanced UPR activation, and that ER-phagy is involved in the selective clearance of ER proteins. As a scaffold protein, the autophagic adapter Ref(2)P/P62 delivers ubiquitinated proteins for selective autophagic degradation by: (1) interacting with LC3/ATG8 through its LIR region, and (2) interacting with ubiquitinated proteins through its ubiquitin-associated (UBA) domain (Moscat and Diaz-Meco, 2012; Nezis et al., 2008). In prolonged ER stress (both Rh1P37H-GFP perkRNAi and ninaEG69D photoreceptor neurons), membrane proteins including rhodopsin get stuck and ubiquitinated in ER. Thus Ref(2)P/P62 and LC3/ATG8 proteins also accumulate, and wild-type rhodopsin and ER-GFP can be detected within Ref(2)P/P62 puncta. By contrast, the mitochondrial reporter mito-GFP did not associate with these puncta. In both Rh1P37H-GFP perkRNAi and ninaEG69D photoreceptor cells, levels of ER-GFP and wild-type rhodopsin were dramatic reduced, whereas proteins in other cellular compartments (mito-GFP and cytosolic GFP) were less affected. When we knocked down ref(2)P/p62 or autophagy-relative genes, this UPR induced autophagy was disrupted and degradation of wild-type rhodopsin was prevented. Our results suggested that tuning down selective autophagy may protect against retinal degeneration. Interestingly, it has recently been reported that disrupting the interaction between Ref(2)P/P62 and Atg8a increases the tolerance to oxidative stress and reduces levels of aging-associated mitochondrial superoxide in Drosophila (Bhattacharjee et al., 2022).
Despite being caused by different insults, misfolded membrane proteins accumulated in the ER and were removed through ERAD via ubiquitination. The accumulation of misfolded proteins could exert a dominant negative effect on wild-type ER proteins, as illustrated by the dominant mutations in opsin genes (Colley et al., 1995; Mendes and Cheetham, 2008; Rajan and Kopito, 2005; Saliba et al., 2002). By conducting a genome-wide loss of function screen, PERK and eIF2Bα were found to play a key role in maintaining levels of wild-type rhodopsin in heterozygous Rh1P37H mutants. Further, disruption of the selective autophagy through inactivation of Ref(2)P/P62 prevented the dominant effects of Rh1P37H , and maintained normal levels of wild-type Rh1. Although it has been reported that misfolded CFTR (Cystic Fibrosis Transmembrane Conductance Regulator), and gonadotropin-releasing hormone receptor (GnRHR), are resistant to ERAD and cleared by an ER-associated autophagy (He et al., 2021; Houck et al., 2014), ER-associated autophagy had little effect on the clearance of mutant rhodopsin. Elevated ER-phagy was associated with the accumulation of Rh1P37H, and interruption of autophagy had no effect on Rh1P37H accumulation. This is consistent with results seen in mice that inhibiting autophagy reduced the cytotoxicity of misfolded RhoP23H (Yao et al., 2018). Studies in both Drosophila and mice have demonstrated that increasing ERAD by either induction of E3 ligases or proteasome activity reduced mutated rhodopsin and is cytoprotective (Kang and Ryoo, 2009; Lobanova et al., 2018; Xu et al., 2020). Indeed, blocking PERK signaling resulted in the accumulation of ubiquitinated proteins, preventing the proteasome from degrading its normal target such as GFP-Flag-Cl1. This increased burden on the proteasomal degradation machinery has been implicated in the pathology of multiple inherited retinal degeneration diseases (Lobanova et al., 2013). Specific ubiquitination of Rh1P37H by the E3 ligase SORDD1 dramatically decreased the accumulation of misfolded Rh1P37H in Rh1P37H-GFP perkRNAi photoreceptor cells, but still reduced wild-type Rh1. On the contrary, blocking ER-phagy restored wild-type Rh1 but did not affect Rh1P37H, supporting the notion that Rh1P37H and wild-type Rh1 are cleared by different degradation machineries. Moreover, expressing the deubiquitinase USP15-31, which largely alleviated levels of cytosolic ubiquitination but did not affect ubiquitination of Rh1P37H, prevented the ER-phagic degradation of wild-type Rh1 but not the accumulation of Rh1P37H. The accumulation of ubiquitinated proteins upon proteasome overload in prolonged ER stress, along with upregulation of Ref(2)P/P62, promoted the selective autophagy of general ER proteins including wild-type rhodopsin, whereas unfolded Rh1P37H was not a substrate of this selective autophagy. Rh1P37H may be segregated into a distinct ER substructure. The observation that only wild-type Rh1 but not Rh1P37H was detected in Ref(2)P/P62 puncta further supports this.
Impaired processing of ubiquitinated proteins has been reported in several mouse models of retinal degeneration (Lobanova et al., 2018; Lobanova et al., 2013), as we found in Rh1P37H-GFP perkRNAi photoreceptor cells. Moreover, photoreceptor degeneration in RhoP23H/+ mice can be delayed by increasing photoreceptor proteasomal activity through overexpression of the 11S proteasome cap subunit, PA28α or activation of mTORC1 (Lobanova et al., 2018; Wang et al., 2022). Ubiquitin ligases and the upstream ERAD proteins were upregulated through IRE1/XBP1 signaling, which could induce proteasomal overload. This global impairment of proteasomal function precedes photoreceptor cell death (Chiang et al., 2015). As a result of insufficiency of the UPS system, autophagy is elevated to complement proteasome-dependent degradation. The continuously activating IRE1/XBP1 signaling upregulated multiple autophagy-related genes in Rh1P37H-GFP perkRNAi photoreceptor cells, while in combination with loss of translational inhibition through PERK/eIF2α deficiency, ubiquitinated proteins accumulated. These signals ultimately led to the induction of ubiquitination/P62-mediated selective autophagy. However, the autophagy could not substitute for the UPS system, as Rh1P37H was resistant to autophagy, whereas ER proteins and wild-type Rh1 were found in autophagosomes. This ER-phagy degradation of wild-type Rh1 accelerated the retinal degeneration induced by Rh1P37H. In the fly ninaEG69D model of adRP, prolonged activation of IRE1/XBP1 signaling was associated with autophagy induction, degradation of normal ER proteins, dysfunction in phototransduction, and retinal degeneration, whereas overexpression of PERK largely suppressed these phenotypes. Supporting this point, xbp1 deficiency protects against neurodegeneration in transgenic mouse models of Huntington disease (HD) and amyotrophic lateral sclerosis (ALS), likely through the enhancement of autophagy (Hetz et al., 2009; Vidal et al., 2012).
Under ER stress, PERK is activated by oligomerization and autophosphorylation, which enables the phosphorylation of eIF2α, reducing global translational efficiency by blocking 80S ribosome assembly (Harding et al., 1999; Shi et al., 1998). In addition to eIF2α, activated PERK also phosphorylates and activates other targets such as Nrf2 and FOXO, regulating cellular metabolic adaptation and survival in the context of ER stress (Cullinan et al., 2003; Zhang et al., 2013). When ER stress was induced by misfolded rhodopsin Rh1P37H, PERK-eIF2α signaling is key for maintaining rhodopsin homeostasis. Rhodopsin homeostasis was disrupted by mutations in eIF2Bα, which regulates eIF2α, as well as in perk mutants under mild ER-stress. Expressing the dominant negative form of eIF2α mimicked the phenotype of perk mutant in terms of both accumulation of Rh137H and reduction of wild-type Rh1. Moreover, inhibiting global translation by knocking down eIF3b and eIF4G abolished the effects of perk deficiency on both mutant and wild-type Rh1. Besides reducing global protein biosynthesis, PERK-mediated phosphorylation of eIF2α increases the translation of ATF4 by bypassing upstream open reading frames (uORFs; Harding et al., 2000a). As a transcription factor that activates UPR target genes associated with protein folding and apoptosis, ATF4 plays an essential role in PERK-mediated UPR (Costa-Mattioli and Walter, 2020). However, ATF4 plays a minor role in maintaining homeostasis of rhodopsin through PERK signaling. First, unlike mutations in perk or eIF2Bα, animals with complete loss of atf4 exhibited little changes in both mutant and wild-type rhodopsin. Second, mimicking the increased translation of ATF4 by activation of ATF4 through deletion of uORF did not suppress perk mutation-induced accumulation of Rh137H and reduction of wild-type Rh1. Finally, expression of ATF4 had no effect on the severity of retinal degeneration in the ninaEG69D model, while over-expression of PERK strongly suppressed this retinal degeneration. Besides ATF4, several factors are translationally induced by eIF2α in parallel to ATF4 in both mammalian cells and flies (Andreev et al., 2015; Baird et al., 2014; Brown et al., 2021; Zhou et al., 2008). The fact that translational inhibition by interfering with eIF3b and eIF4G could mimic rhodopsin level phenotypes strongly argues against the possibility that eIF2α-dependent translational induction is required in this PERK signaling.
Protein misfolding and activation of the UPR is emerging as a common mechanism in neurodegeneration diseases. Studies inhibiting the PERK branch of UPR have shown that PERK inhibitors protect against neuronal cell death in models of Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS; Celardo et al., 2016; Kim et al., 2013; Moreno et al., 2013). A protective role of PERK in Charcot-Marie-Tooth (CMT) disease has also been demonstrated, as genetic and pharmacological inactivation of Gadd34 (damage-inducible protein 34) antagonizes PERK activity toward eIF2α and improves motor, neurophysiological, and morphologic deficits in mouse models (Scapin et al., 2020). In mouse models of adRP (heterozygous RhoP23H), PERK protects against degeneration of photoreceptor cells (Athanasiou et al., 2017; Chiang et al., 2012a). Consistent with this, loss-of-function perk mutations dramatically accelerate retinal degeneration in fly ninaEG69D models (Vasudevan et al., 2020). In contrast, limited ATF4 expression protects photoreceptor cells from RhoP23H-induced retinal degeneration, while conflicting evidence suggests that loss of ATF4 accelerates retinal degeneration in Drosophila ninaEG69D models (Bhootada et al., 2016; Vasudevan et al., 2022). Supporting the protective role of PERK but not ATF4 in adRPs, loss of function of PERK led to severe retinal degeneration in Rh1P37H-expressing cells, whereas a null ATF4 allele did not. Importantly, overexpression of PERK strongly suppressed retinal degeneration in ninaEG69D flies, whereas ATF4 had no effect. In accordance with the deteriorating role of autophagy, the degenerating ninaEG69D flies exhibit increases in autophagy (in particular ER-phagy) and activation of PERK prevents autophagy, suggesting that reduced autophagy flux is part of the protective mechanism of PERK (Qiu et al., 2019; Yao et al., 2018). Our findings indicate that prolonging PERK activities represent a valid therapeutic target in ER stress-related neuropathies such as adRP.
Materials and methods
Fly stocks
The following fly lines were obtained from the Bloomington Stock Center: P{UAS-LUC.VALIUM10}attP2 (LUCRNAi), P{TRiP.HMS03003}attP40 (ire1RNAi-1), P{TRiP.HMC05163}attP40 (ire1RNAi-2), P{TRiP.HMS03015}attP2 (xbp1RNAi-1), P{TRiP.JF02012}attP2 (xbp1RNAi-2), w1118;PBac{w[+mC] = WH}ire1[f02170]/TM6B,Tb (ire1f02170), P{TRiP.HMJ02063}attP40 (perkRNAi). The following fly lines were obtained from the Tsinghua Fly Center: P{TRiP. HMS00668} attP2 (eIF3bRNAi), P{TRiP. HMS00762} attP2 (eIF4GRNAi), P{TRiP. HMS02750} attP40 (atg1RNAi), P{TRiP. JF02891} attP2 (atg9RNAi), P{TRiP. HMS01193} attP2 (atg18RNAi), P{TRiP. HMS00551} attP2 (ref(2)P/p62RNAi-1), P{TRiP. HMS00938} attP2 (ref(2)P/p62RNAi-2), P{TRiP.HMS00139} attP2 (prosβ1RNAi). The knock-down efficiency of RNAi lines was verified by qPCR. The following flies were maintained in the laboratory of T. Wang: ATF4-mCherry, ey-flp; FRT82B GMR-hid CL/TM3, ninaE-GFP, trp-GFP, GMR-Gal4, ninaE-xbp1-RE. All flies were maintained under 12 h light/12 h dark cycles at 25°C unless mentioned.
Generation of transgenic flies
The cDNA sequences of Rh1, perk, eIF2Bα, atf4, and eIF2α were amplified from RH01460, LD41715, HL01112, RH01327, and GH06180 of the DGRC gold cDNA collections, respectively (Drosophila Genomics Resource Center). The Rh1 cDNA was subcloned into the pninaE-attB vector with a C-terminal RFP-tag. Rh1P37H was mutated from the ninaE cDNA and subcloned into the pninaE-attB vector with a C-terminal GFP-tag. The perk cDNA was controlled by its own promotor and subcloned into the Pperk-attB vector. The eIF2Bα cDNA was subcloned into a ubi-attB vector. The atf4 cDNA was subcloned into a da-attB vector. The eIF2α cDNA and eIF2ΑS51A mutated from eIF2α cDNA were subcloned into a pUAST-attB vector. The mCherry-T2A-GFP-FLAG-Cl1 sequence was subcloned into the pninaE-attB vector. The constructs were injected into M (vas-int.Dm) ZH-2A;M(3xP3-RFP.attP)ZH-86Fb or M (vas-int.Dm) ZH-2A;M (3xP3-RFP.attP) ZH-51C embryos, and transformants were identified based on eye color. The 3XP3-RFP markers were eliminated by crossing to a Cre-expressing line. To generate the xbp1-mCherry reporter flies, the EGFP of P{UAS-Xbp1.EGFP.HG} (Ryoo et al., 2007) was replaced by mCherry.
Generation of atf4, atf6, and ref(2)P knockout flies (atf4KO, atf61, and ref(2)Pm)
The atf4KO mutation was generated using the Cas9/sgRNA system (Fig. S4 A; Xu, 2015). Briefly, a pair of guide RNAs targeting the atf4 locus were designed (sgRNA1: 5′-CTGATTACCAGCTCAATGAT-3′, sgRNA2: 5′-ACGAGGACTGGGTTCCAGAG-3′) and cloned into the U6b-sgRNA-short vector. Plasmids were injected into the embryos of nos-Cas9 flies, and deletions were identified by PCR using the following primers: forward primer 5′-AAATTGTTTGGCCTCTTTGATG-3′ and reverse primer 5′-TTTTCCTGATCTTTCGATCCTC-3′.
The atf61 mutation was generated through a combination of sgRNA targeting and Flp/FRT recombination as shown in Fig S4 C. Briefly, a pair of guide RNAs targeting the atf6 locus were designed (sgRNA1: 5′-AATAGAAGGCTCGTGTCGGT-3′, sgRNA2: 5′-TCTTGTTGTACAGTATTGAC-3′) and cloned into the U6b-sgRNA-short vector. A pair of homology arm (800 bp upstream and 800 bp downstream of each sgRNA site) sequences were cloned into the 5′ and 3′ ends of a FRT sequence (5′-GAAGTTCCTATACTTTCTAGAGAATAGGAACTTC-3′). The two pairs of sgDNA and FRT constructs were injected into the embryos of nos-Cas9 flies, and the flies with both FRT sites replaced were identified by PCR using the following primers: FRT-up: 5′-CAATAGAAGGGAAGTTCCTATA-3′; FRT-down: 5′-CTGTCACATACCTGAAGTTCCT-3′.
The atf6FRT knock-in flies, were cross with hs-flp lines to delete the DNA fragments between the two FRT sites. The following PCR primers were used to verify the atf61 flies: ATF6-FRT del-F: 5′-CGCTAACCTCAATACGAAATGG-3′; ATF6-FRT del-R: 5′-TTCATACGGACAGACGGACATA-3′.
The ref(2)Pm mutation was generated using a single guide DNA (sgDNA: 5′-ACTGAGTCAAGACTCCGGCA-3′). The plasmid was injected into nos-Cas9 embryos. A mutant allele in which 4 nucleotides were deleted was identified by PCR and DNA sequencing using the following primers: 5′-f-GAGCCCTCAGTGATTCACCT-3′ and 5′-r-CTGGATTGACCCTGCTCTTCT-3′. All mutant flies generated were backcrossed to wild-type flies (w1118) before preforming experiment.
EMS mutagenesis
The second chromosome of FRT40A;Rh1P37H-GFP or FRT42D;Rh1P37H-GFP flies and the third chromosome of FRT2A Rh1P37H-GFP or FTR82B Rh1P37H-GFP flies were isogenized, and young male flies were fed 25 mM EMS (Sigma-Aldrich) in 2% sucrose for 8 h, followed by mating to ey-flp Rh1-RFP;FRT40A GMR-hid CL/Cyo, ey-flp Rh1-RFP;FRT42D GMR-hid CL/Cyo, ey-flp Rh1-RFP;FRT2A GMR-hid CL/TM3 and ey-flp Rh1-RFP;FRT82B GMR-hid CL/TM3 flies, respectively. Approximately 100,000 F1 progeny for each chromosomal arm were examined for fluorescence of GFP-tagged Rh1P37H and RFP-tagged wild-type Rh1 (Fig S2A).
Fly imaging
Flies were anaesthetized by CO2, and fluorescence images were taken with a Leica M165 FC Fluorescent Stereo Microscope.
Immunoprecipitation and Western blots
Approximately 200 fly heads were lysed with 10 mM Tris-HCl lysis buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40, 0.09% Na-Azide with 1× proteinase inhibitor cocktail [Roche] and 1 mM PMSF) for 30 min, and centrifuged at 20,000 g. The supernatant was used for subsequent immunoprecipitation with anti-GFP beads (Chromotek). Beads were washed in ice-cold dilution buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA, 0.018% Na-Azide with 1× proteinase inhibitor cocktail and 1 mM PMSF) three times and boiled (45°C, 700 rpm, 30 min) in SDS loading buffer for standard Western blot assays.
For Western blot assays, dissected adult fly heads were homogenized with a pellet pestle (Thermo Fisher Scientific) in SDS loading buffer for SDS-PAGE. The blots were probed with primary antibodies against Rh1 (mouse, 1:2,000; Developmental Studies Hybridoma Bank), INAD (rat, 1:2,000; Wang et al., 2008), mouse anti-β-actin (1:2,000; Santa Cruz), mouse anti-ubiquitin (1:1,000; Santa Cruz), rabbit anti-GFP (1:3,000; easybio), tubulin (mouse, 1:2,000; Developmental Studies Hybridoma Bank), RFP (rabbit, 1:2,000; Biovision), TRP (rabbit, 1:2,000; Wang et al., 2008), Flag (mouse, 1:2,000; Sigma-Aldrich), Ref(2)P/P62 (rabbit, 1:2,000; Abmart), followed by incubation with IRDye 680 goat anti-mouse IgG (1:10,000; LI-COR Biosciences), IRDye 800 goat anti-rabbit IgG (1:10,000, LI-COR Biosciences), or IRDye 680 goat anti-rat IgG (1:10,000; LI-COR Biosciences). Signals were detected using an Odyssey infrared imaging system (LI-COR Biosciences).
Immunostaining
Fly heads were cut and fixed in 4% freshly made paraformaldehyde (Sigma-Aldrich) in phosphate buffer for 2 h on ice, then the retinas were dissected for immunostaining. Samples were incubated with primary antibodies including mouse anti-Rh1 (1:200; Developmental Studies Hybridoma Bank), rat anti–INAD (1:200; Wang et al., 2008), rat anti–RFP antibody (1:200; Chromotek), rabbit anti-Ref(2)P/P62 (1:200; Abmart), rabbit anti-calnexin (1:200; Zhao and Wang, 2020), rabbit anti-Atg8a (1:200; Abcam) at 4 °C overnight, followed by incubation with Alexa Fluor 488–conjugated, Alexa Fluor 568–conjugated, Alexa Fluor 647–conjugated secondary antibodies (1:500; Invitrogen), and 20 nM DAPI (Invitrogen) or phalloidin (Invitrogen) for 1 h at room temperature. Fluorescence images were acquired at room temperature using a LSM800 confocal laser scanning microscope (Zeiss) with a 63×/1.4-NA oil-immersion lens or a 40×/0.95-NA dry lens and Zeiss Application Suite ZEN software (Zeiss Microsystems).
ERG recordings
ERG recordings were performed as described (Wang et al., 2008). Briefly, two glass microelectrodes filled with Ringer’s solution were placed on small drops of electrode cream (Parker Laboratories) on the compound eye and the thorax of a fly. For PDA recordings, flies were dark adapted for 2 min. White-eyed flies were then exposed to 5 s of orange light, followed by two rounds of 5 s of blue light, and two rounds of 5 s of orange light. The time between two stimulations was 5 s. For the summed light responses of photoreceptor cells, flies were dark adapted for 2 min and then white-eyed flies were exposed to a 1-s pulse of ∼2,000 lux orange light (source light was filtered using an FSR-OG550 filter, Newport). ERG signals were amplified with a Warner electrometer IE-210 and recorded with a MacLab/4 s analog-to-digital converter and the clampelx 10.2 program (Warner Instruments). All recordings were carried out at room temperature.
Transmission electron microscopy
TEM was performed with standard methods as described (Xu and Wang, 2016). Briefly, fly heads were cut and fixed in 4% paraformaldehyde and 2.5% glutaraldehyde at 4°C overnight followed by incubation in 1% osmium tetroxide for 1–2 h at 4°C. Then the samples were dehydrated using a series of ethanol dilutions (10, 25, 35, 40, 55, 70, 85, 95, and 100% ethanol) and embedded in LR White resin (Polysciences, Inc.). Thin sections (80 nm) were stained with uranyl acetate and lead citrate (Sigma-Aldrich) and examined using a JEOL JEM-1400 transmission electron microscope (JEOL Ltd.) at room temperature. The images were acquired using a Gatan CCD (4 k × 3.7 k pixels).
RNA extraction and quantitative real-time PCR analysis
Fly eyes were dissected, and RNA was extracted using TRIzol Reagent (Invitrogen). Total RNA was reverse-transcribed using EasyScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen). qRT-PCR was performed using iTaq Universal SYBR Green Supermix (BIO-RAD) on a CFX96 real time PCR detection system (Bio-Rad). The average threshold cycle value (CT) was calculated from at least three replicates per sample. Expression of genes were standardized relative to rp49. Relative expression values were determined by the ΔΔ CT method. Primers used as below.
rp49-F: 5′-AGCATACAGGCCCAAGATCG-3′, rp49-R: 5′-TGTTGTCGATACCCTTGGGC-3′
Hsc70-3-F: 5′-GGTAACCGCATCACTCCCTC-3′, Hsc70-3-R: 5′-GTGGTCAACTGATTCTTGGCG-3′
BI-1-F: 5′-GCCACTCTAGTCCTGGTCTTG-3′, BI-1-R: 5′-GCCGGAGCAGAATCCGAAG-3′
Gp93-F: 5′-ATCCGCCTATTGGCTCTGTC-3′, Gp93-R: 5′-CCGAGTCCATGATGTGCAAC-3′
CaBP1-F: 5′-GAGGTGCTGAAAGACGACG-3′, CaBP1-R: 5′-CGACTCCCTTCAATGCCTTGG-3′
Ero1L-F: 5′-CTTCTTCCGCTTCTACAAGGTG-3′, Ero1L-R: 5′-CTTGATGCCCTGGGGAATCG-3′
-
Sec22-F: 5′-GGACGCAGCATACTGGACTAC-3′, Sec22-R: 5′-GTCCGGTCTCGATACTGCATC-3′
atg1-F: 5′-CGTCAGCCTGGTCATGGAGTA-3′, atg1-R: 5′-TAACGGTATCCTCGCTGAG-3′
atg2-F: 5′-ATGCGCTGATGACCAACGA-3′, atg2-R: 5′-CCGACGACCACATGGACTC-3′
atg3-F: 5′-TCAATGTGGCCGAATATCTGAC-3′, atg3-R: 5′-AGGTAGGGTTTTGTCTTGGTCT-3′
atg8a-F: 5′-GGTCAGTTCTACTTCCTCATTCG-3′, atg8a-R: 5′-GATGTTCCTGGTACAGGGAGC-3′
atg9-F: 5′-TCTAGCCCACATATCAACTACCG-3′, atg9-R: 5′-CTTTTGCGTCTTGTGTTTTGGAT-3′
atg18a-F: 5′-ACCACACGAAAAGCGACGAG-3′, atg18a-R: 5′-5′-GCTCTGCTTCTTAAAGTGGCAC-3′
perk-F: 5′-TACTAGGTCCAGTGGTGC-3′, perk-R: 5′-GCTTGTCCAGGTGGGAAGCTA-3′
ire1-F: 5′-ACTTCGCGGGCCATCTATCTA-3′, ire1-R: 5′-GCACTCACAGCATTGTAGTCGTA-3′
eIF3B-F: 5′-GGATGCGAACGACAGTGATTA-3′, eIF3B-R: 5′-GGGATATTGTCCACTACCACCA-3′
prosβ1-F: 5′-GGTCATTGGAGCCGATTCG-3′, prosβ1-R: 5′-GCAGTACACTTTGTCCGTGAT-3′
xbp1-F: 5′-CCGAACTGAAGCAGCAACAGC-3′, xbp1-R: 5′-CAGAGGGTCAGCTTTGGATGC-3′
xbp1-splicing-F: 5′-CCGAACTGAAGCAGCAACAGC-3′,
xbp1-splicing-R: 5′-ATACCCTGCGGCAGATCCAA-3′
ref(2)P/p62-F: 5′-AATCGAGCTGTATCTTTTCCAGG-3′,
ref(2)P/p62-R: 5′-AACGTGCATATTGCTCTCGCA-3′
eIF4G-F: 5′-TATAACCCACGGCAACAAACAT-3′,
eIF4G-R: 5′-TGCTGAAGAGTTGGGACATATTG-3′.
RNA sequencing
RNA sequencing (RNA-seq) assays were performed to analyze the transcriptome of fly retinas. Briefly, 20 retinas were dissected from 1-d-old flies. Total RNA was purified using Trizol reagent. RNA integrity was checked using a 2,100 Bioanalyzer (Agilent Technologies) with a minimum RNA integrity number of 8. The mRNA was enriched using oligo magnetic beads (Invitrogen) and fragmented to ∼150–250 bp. cDNAs were synthesized using random hexamer primers and purified using a MinElute PCR purification kit (Qiagen). The 42-cycle single-end sequencing was performed using an Illumina Genome Analyzer IIx. CASAVA pipeline v1.8 was then used for sequence extraction and filtering. RNA-seq reads were mapped to the fly genome using Tophat (v2.0.8b) software and the Ensembl genome annotation dataset (Drosophila_melanogaster.BDGP5.71.gtf). Gene expression level fragments per kilobase of exon per million fragments mapped (FPKM) was estimated using Cufflinks (v2.1.1) software.
Proteomic analysis of the fly retina
40 retina pairs for each genotype (1-d-old) were dissected in cold phosphate buffer. Two samples were generated for each genotype. Protein extraction was performed as described using 50 μl lysis buffer (10% SDS with 100 mM TEAB) for 30 mins on ice and centrifuged at 16,000 g to collect supernatant (Xiong et al., 2020). About 50 μg protein for each sample was collected and digested with trypsin (Promega Corporation) in an enzyme/protein ratio of 1:50 (w/w) overnight at 37°C. The resulting peptide samples were labeled using the TMT 10plex Isobaric Label Reagent Set label kit (Thermo). The mixed peptides were fractionated using a reversed phase C18 column (3 M, Bracknell), and 8 fractions of peptide were eluted with acetonitrile step gradients (7.5, 10, 12.5, 15, 17.5, 20, 22.5, and 50%, pH 10). Finally, the eight fractions were dried in a vacuum centrifuge and stored at −80°C until LCMS/MS analysis.
Approximately 2 μg of each pH fractionated peptide sample were separated on an in-house packed 75-μm ID × 50 cm capillary column with 2.5 μm Venusil C18 beads (Agela Technologies) using an EASY-nLC 1,000 system (Thermo Fisher Scientific) with flow rate at 200 nl/min. Raw data was collected on Q Exactive mass spectrometer (Thermo Fisher Scientific) using Thermo Xcalibur (2.0) software. All raw LC−MS/MS data were submitted to Proteome Discoverer (2.2 version, Thermo Science) for TMT quantitation and database analysis using SequestHT. Data were searched against the Fruit fly Swiss-Prot database (UP000000803, 21,922 sequences) in combination with a common contaminants database (247 entries).
Statistics
Statistical results were generated by GraphPad Prism 6 and statistical significance was assessed through Ordinary one/two-way ANOVA, Sidak’s multiple comparisons test, and Student’s t test analyses. All error bars represent standard error of the mean. ImageJ was used to quantify the fluorescence intensity of Western blot and immunostaining images. A graphical normality test was performed to determine whether all data used for statistical analysis were normally distributed. Briefly, a histogram of the dataset was created, and all data fall in a normally distributed population.
Online supplemental material
Fig. S1 shows that a genetic screen reveals that perk and eIF2Bα are involved in Rh1 homeostasis in a model of adRP. Fig. S2 shows the levels of Rh1P37H and endogenous Rh1 are unaffected by mutations in ire1 and atf6. Fig. S3 shows the overlap of upregulated genes in Rh1P37H-GFP perkRNAi and ninaE-xbp1-RE retinas. Fig. S4 shows that the ER proteins are degraded through autophagy in Rh1P37H-GFP perkRNAi photoreceptor cells. Fig. S5 shows that the ER-phagy is induced in the ninaEG69D model of adRP. Data S1 shows the gene expression profiling of retina of the Rh1P37H-GFP perkRNAi and Rh1P37H-GFP flies related to Fig. 4 A. Data S2 shows the gene expression profiling of retina dissected from the ninaE-GFP and ninaE-xbp1-RE flies related to Fig. S3 A. Data S3 shows comparison of retinal protein levels of Rh1P37H-GFP perkRNAi with Rh1P37H-GFP flies related to Fig. 5 A.
Supplementary Material
shows the gene expression profiling of retina of the Rh1P37H-GFP/perkRNAi and Rh1P37H-GFP flies related to Fig. 4 A.
shows the gene expression profiling of retina dissected from the ninaE-GFP and ninaE-xbp1-RE flies related to Fig. S3 A.
shows comparison of retinal protein levels of Rh1P37H-GFP perkRNAi with Rh1P37H-GFP flies related to Fig. 5 A.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S2.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We thank the Bloomington Stock Center, the Drosophila Genomic Resource Center, the Tsinghua Fly Center, and the Developmental Studies Hybridoma Bank for stocks and reagents. We thank Y. Wang and X. Liu for assistance with fly injections. We are tremendously thankful for support provided by the Sequencing Center, the Electron Microscopy Center, the Imaging Facility, and the Proteomics Facility at the National Institute of Biological Sciences. We thank Dr. Lin Yang from the Institute of Genetics and Developmental Biology for assistance with TEM. We thank Dr. D. O’Keefe for editing the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (81870693 and 81670891) awarded to T. Wang.
Author contributions: Methodology: T. Wang, N. Zhao. Investigation: N. Zhao. Visualization: N. Zhao, N. Li. Supervision: TW. Writing—original draft: T. Wang, N. Zhao. Writing—review & editing: T. Wang, N. Zhao.
Data availability
All data are available in the main text or the supplementary materials.
References
- Adomavicius, T., Guaita M., Zhou Y., Jennings M.D., Latif Z., Roseman A.M., and Pavitt G.D.. 2019. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10:2136. 10.1038/s41467-019-10167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., and Harper J.W.. 2019. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell. 74:891–908.e10. 10.1016/j.molcel.2019.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev, D.E., O’Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., and Baranov P.V.. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 4:e03971. 10.7554/eLife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou, D., Aguila M., Bellingham J., Kanuga N., Adamson P., and Cheetham M.E.. 2017. The role of the ER stress-response protein PERK in rhodopsin retinitis pigmentosa. Hum. Mol. Genet. 26:4896–4905. 10.1093/hmg/ddx370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou, D., Aguila M., Bellingham J., Li W., McCulley C., Reeves P.J., and Cheetham M.E.. 2018. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 62:1–23. 10.1016/j.preteyeres.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- B’chir, W., Maurin A.C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., and Bruhat A.. 2013. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41:7683–7699. 10.1093/nar/gkt563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, T.D., Palam L.R., Fusakio M.E., Willy J.A., Davis C.M., McClintick J.N., Anthony T.G., and Wek R.C.. 2014. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell. 25:1686–1697. 10.1091/mbc.e14-02-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, D.L., and Baehrecke E.H.. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148. 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, A., Ürmösi A., Jipa A., Kovács L., Deák P., Szabó Á., and Juhász G.. 2022. Loss of ubiquitinated protein autophagy is compensated by persistent cnc/NFE2L2/Nrf2 antioxidant responses. Autophagy. 18:2385–2396. 10.1080/15548627.2022.2037852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhootada, Y., Kotla P., Zolotukhin S., Gorbatyuk O., Bebok Z., Athar M., and Gorbatyuk M.. 2016. Limited ATF4 expression in degenerating retinas with ongoing ER stress promotes photoreceptor survival in a mouse model of autosomal dominant retinitis pigmentosa. PLoS One. 11:e0154779. 10.1371/journal.pone.0154779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, B., Mitra S., Roach F.D., Vasudevan D., and Ryoo H.D.. 2021. The transcription factor Xrp1 is required for PERK-mediated antioxidant gene induction in Drosophila. Elife. 10:e74047. 10.7554/eLife.74047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon, M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., and Ron D.. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Celardo, I., Costa A.C., Lehmann S., Jones C., Wood N., Mencacci N.E., Mallucci G.R., Loh S.H., and Martins L.M.. 2016. Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell Death Dis. 7:e2271. 10.1038/cddis.2016.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, T.K., Lawrence D.A., Lu M., Tan J., Harnoss J.M., Marsters S.A., Liu P., Sandoval W., Martin S.E., and Ashkenazi A.. 2018. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol. Cell. 71:629–636.e5. 10.1016/j.molcel.2018.06.038 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Xiao Y., Chai P., Zheng P., Teng J., and Chen J.. 2019. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 29:846–855.e6. 10.1016/j.cub.2019.01.041 [DOI] [PubMed] [Google Scholar]
- Chiang, W.C., Hiramatsu N., Messah C., Kroeger H., and Lin J.H.. 2012a. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest. Ophthalmol. Vis. Sci. 53:7159–7166. 10.1167/iovs.12-10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, W.C., Kroeger H., Sakami S., Messah C., Yasumura D., Matthes M.T., Coppinger J.A., Palczewski K., LaVail M.M., and Lin J.H.. 2015. Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Mol. Neurobiol. 52:679–695. 10.1007/s12035-014-8881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, W.C., Messah C., and Lin J.H.. 2012b. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol. Biol. Cell. 23:758–770. 10.1091/mbc.e11-08-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, D.S., Cairrão F., Zeng X., Pires E., Coelho A.V., Ron D., Ryoo H.D., and Domingos P.M.. 2013. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep. 5:791–801. 10.1016/j.celrep.2013.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley, N.J., Cassill J.A., Baker E.K., and Zuker C.S.. 1995. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc. Natl. Acad. Sci. USA. 92:3070–3074. 10.1073/pnas.92.7.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli, M., and Walter P.. 2020. The integrated stress response: From mechanism to disease. Science. 368:368. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J.S., Shamu C.E., and Walter P.. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. 10.1016/0092-8674(93)90648-A [DOI] [PubMed] [Google Scholar]
- Cullinan, S.B., Zhang D., Hannink M., Arvisais E., Kaufman R.J., and Diehl J.A.. 2003. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23:7198–7209. 10.1128/MCB.23.20.7198-7209.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, J., and Baehrecke E.H.. 2018. Life, death and autophagy. Nat. Cell Biol. 20:1110–1117. 10.1038/s41556-018-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja, T.P., McGee T.L., Reichel E., Hahn L.B., Cowley G.S., Yandell D.W., Sandberg M.A., and Berson E.L.. 1990. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 343:364–366. 10.1038/343364a0 [DOI] [PubMed] [Google Scholar]
- Fawcett, T.W., Martindale J.L., Guyton K.Z., Hai T., and Holbrook N.J.. 1999. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339:135–141. 10.1042/bj3390135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli, F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., Fregno I., Galli C., Loi M., Soldà T., et al. 2016. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 18:1173–1184. 10.1038/ncb3423 [DOI] [PubMed] [Google Scholar]
- Galy, A., Roux M.J., Sahel J.A., Léveillard T., and Giangrande A.. 2005. Rhodopsin maturation defects induce photoreceptor death by apoptosis: A fly model for RhodopsinPro23His human retinitis pigmentosa. Hum. Mol. Genet. 14:2547–2557. 10.1093/hmg/ddi258 [DOI] [PubMed] [Google Scholar]
- Gilon, T., Chomsky O., and Kulka R.G.. 1998. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17:2759–2766. 10.1093/emboj/17.10.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc, A., Aron L., Roux M.J., Klein R., Giangrande A., and Ueffing M.. 2010. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 6:e1001075. 10.1371/journal.pgen.1001075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati, P., Morozzi G., Hölper S., Mari M., Harwardt M.I., Yan R., Müller S., Reggiori F., Heilemann M., and Dikic I.. 2017. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 6:e25555. 10.7554/eLife.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., and Ron D.. 2000a. a. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang Y., Bertolotti A., Zeng H., and Ron D.. 2000b. b. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang Y., and Ron D.. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 397:271–274. 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Hartong, D.T., Berson E.L., and Dryja T.P.. 2006. Retinitis pigmentosa. Lancet. 368:1795–1809. 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- Haze, K., Yoshida H., Yanagi H., Yura T., and Mori K.. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 10:3787–3799. 10.1091/mbc.10.11.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Kennedy A.S., Houck S., Aleksandrov A., Quinney N.L., Cyr-Scully A., Cholon D.M., Gentzsch M., Randell S.H., Ren H.Y., and Cyr D.M.. 2021. DNAJB12 and Hsp70 triage arrested intermediates of N1303K-CFTR for endoplasmic reticulum-associated autophagy. Mol. Biol. Cell. 32:538–553. 10.1091/mbc.E20-11-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz, C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., Martinez G., Cuervo A.M., Brown R.H., and Glimcher L.H.. 2009. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23:2294–2306. 10.1101/gad.1830709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz, C., Zhang K., and Kaufman R.J.. 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21:421–438. 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien, J., and Weissman J.S.. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 313:104–107. 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Houck, S.A., Ren H.Y., Madden V.J., Bonner J.N., Conlin M.P., Janovick J.A., Conn P.M., and Cyr D.M.. 2014. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol. Cell. 54:166–179. 10.1016/j.molcel.2014.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, M.J., and Ryoo H.D.. 2009. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl. Acad. Sci. USA. 106:17043–17048. 10.1073/pnas.0905566106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner, L.R., Anand A.A., Nguyen H.C., Myasnikov A.G., Klose C.J., McGeever L.A., Tsai J.C., Miller-Vedam L.E., Walter P., and Frost A.. 2019. eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science. 364:491–495. 10.1126/science.aaw2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets, A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 522:354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Kim, K.H., Jeong Y.T., Kim S.H., Jung H.S., Park K.S., Lee H.Y., and Lee M.S.. 2013. Metformin-induced inhibition of the mitochondrial respiratory chain increases FGF21 expression via ATF4 activation. Biochem. Biophys. Res. Commun. 440:76–81. 10.1016/j.bbrc.2013.09.026 [DOI] [PubMed] [Google Scholar]
- Klaips, C.L., Jayaraj G.G., and Hartl F.U.. 2018. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217:51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K.. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Kurada, P., and O’Tousa J.E.. 1995. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 14:571–579. 10.1016/0896-6273(95)90313-5 [DOI] [PubMed] [Google Scholar]
- Kurtishi, A., Rosen B., Patil K.S., Alves G.W., and Møller S.G.. 2019. Cellular proteostasis in neurodegeneration. Mol. Neurobiol. 56:3676–3689. 10.1007/s12035-018-1334-z [DOI] [PubMed] [Google Scholar]
- Lee, E.J., Chan P., Chea L., Kim K., Kaufman R.J., and Lin J.H.. 2021. ATF6 is required for efficient rhodopsin clearance and retinal homeostasis in the P23H rho retinitis pigmentosa mouse model. Sci. Rep. 11:16356. 10.1038/s41598-021-95895-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., and Walter P.. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science. 318:944–949. 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.Y., Schröder M., and Kaufman R.J.. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275:24881–24885. 10.1074/jbc.M004454200 [DOI] [PubMed] [Google Scholar]
- Lobanova, E.S., Finkelstein S., Li J., Travis A.M., Hao Y., Klingeborn M., Skiba N.P., Deshaies R.J., and Arshavsky V.Y.. 2018. Increased proteasomal activity supports photoreceptor survival in inherited retinal degeneration. Nat. Commun. 9:1738. 10.1038/s41467-018-04117-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanova, E.S., Finkelstein S., Skiba N.P., and Arshavsky V.Y.. 2013. Proteasome overload is a common stress factor in multiple forms of inherited retinal degeneration. Proc. Natl. Acad. Sci. USA. 110:9986–9991. 10.1073/pnas.1305521110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, H.F., and Cheetham M.E.. 2008. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum. Mol. Genet. 17:3043–3054. 10.1093/hmg/ddn202 [DOI] [PubMed] [Google Scholar]
- Mendes, H.F., van der Spuy J., Chapple J.P., and Cheetham M.E.. 2005. Mechanisms of cell death in rhodopsin retinitis pigmentosa: Implications for therapy. Trends Mol. Med. 11:177–185. 10.1016/j.molmed.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Meusser, B., Hirsch C., Jarosch E., and Sommer T.. 2005. Erad: The long road to destruction. Nat. Cell Biol. 7:766–772. 10.1038/ncb0805-766 [DOI] [PubMed] [Google Scholar]
- Moore, K., and Hollien J.. 2015. Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol. Biol. Cell. 26:2873–2884. 10.1091/mbc.E15-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, J.A., Halliday M., Molloy C., Radford H., Verity N., Axten J.M., Ortori C.A., Willis A.E., Fischer P.M., Barrett D.A., and Mallucci G.R.. 2013. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 5:206ra138. 10.1126/scitranslmed.3006767 [DOI] [PubMed] [Google Scholar]
- Mori, K., Ma W., Gething M.J., and Sambrook J.. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 74:743–756. 10.1016/0092-8674(93)90521-Q [DOI] [PubMed] [Google Scholar]
- Moscat, J., and Diaz-Meco M.T.. 2012. p62: A versatile multitasker takes on cancer. Trends Biochem. Sci. 37:230–236. 10.1016/j.tibs.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P., Varga A., Pircs K., Hegedűs K., and Juhász G.. 2013. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 9:e1003664. 10.1371/journal.pgen.1003664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis, I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D., Rusten T.E., Stenmark H., and Brech A.. 2008. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180:1065–1071. 10.1083/jcb.200711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, T., and Hasegawa M.. 2009. A cellular model to monitor proteasome dysfunction by α-synuclein. Biochemistry. 48:8014–8022. 10.1021/bi900619j [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Tousa, J.E., Baehr W., Martin R.L., Hirsh J., Pak W.L., and Applebury M.L.. 1985. The Drosophila ninaE gene encodes an opsin. Cell. 40:839–850. 10.1016/0092-8674(85)90343-5 [DOI] [PubMed] [Google Scholar]
- Ogata, M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., et al. 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26:9220–9231. 10.1128/MCB.01453-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princely Abudu, Y., Pankiv S., Mathai B.J., Håkon Lystad A., Bindesbøll C., Brenne H.B., Yoke Wui Ng M., Thiede B., Yamamoto A., Mutugi Nthiga T., et al. 2019. NIPSNAP1 and NIPSNAP2 act as “eat me” signals for mitophagy. Dev. Cell. 49:509–525.e12. 10.1016/j.devcel.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Qiu, Y., Yao J., Jia L., Thompson D.A., and Zacks D.N.. 2019. Shifting the balance of autophagy and proteasome activation reduces proteotoxic cell death: A novel therapeutic approach for restoring photoreceptor homeostasis. Cell Death Dis. 10:547. 10.1038/s41419-019-1780-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan, R.S., and Kopito R.R.. 2005. Suppression of wild-type rhodopsin maturation by mutants linked to autosomal dominant retinitis pigmentosa. J. Biol. Chem. 280:1284–1291. 10.1074/jbc.M406448200 [DOI] [PubMed] [Google Scholar]
- Rosenbaum, E.E., Vasiljevic E., Brehm K.S., and Colley N.J.. 2014. Mutations in four glycosyl hydrolases reveal a highly coordinated pathway for rhodopsin biosynthesis and N-glycan trimming in Drosophila melanogaster. PLoS Genet. 10:e1004349. 10.1371/journal.pgen.1004349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, H.D., Domingos P.M., Kang M.J., and Steller H.. 2007. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 26:242–252. 10.1038/sj.emboj.7601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami, S., Maeda T., Bereta G., Okano K., Golczak M., Sumaroka A., Roman A.J., Cideciyan A.V., Jacobson S.G., and Palczewski K.. 2011. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J. Biol. Chem. 286:10551–10567. 10.1074/jbc.M110.209759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba, R.S., Munro P.M., Luthert P.J., and Cheetham M.E.. 2002. The cellular fate of mutant rhodopsin: Quality control, degradation and aggresome formation. J. Cell Sci. 115:2907–2918. 10.1242/jcs.115.14.2907 [DOI] [PubMed] [Google Scholar]
- Sano, R., and Reed J.C.. 2013. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta. 1833:3460–3470. 10.1016/j.bbamcr.2013.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin, C., Ferri C., Pettinato E., Bianchi F., Del Carro U., Feltri M.L., Kaufman R.J., Wrabetz L., and D’Antonio M.. 2020. Phosphorylation of eIF2α promotes schwann cell differentiation and myelination in CMT1B mice with activated UPR. J. Neurosci. 40:8174–8187. 10.1523/JNEUROSCI.0957-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu, C.E., and Walter P.. 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028–3039. 10.1002/j.1460-2075.1996.tb00666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Chen X., Hendershot L., and Prywes R.. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 3:99–111. 10.1016/S1534-5807(02)00203-4 [DOI] [PubMed] [Google Scholar]
- Shi, Y., Vattem K.M., Sood R., An J., Liang J., Stramm L., and Wek R.C.. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499–7509. 10.1128/MCB.18.12.7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders, M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., Yates J.R. III, Su A.I., Kelly J.W., and Wiseman R.L.. 2013. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3:1279–1292. 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, D., Katow H., Huang H.W., Tang G., and Ryoo H.D.. 2022. A protein-trap allele reveals roles for Drosophila ATF4 in photoreceptor degeneration, oogenesis and wing development. Dis. Model. Mech. 15:dmm049119. 10.1242/dmm.049119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, D., Neuman S.D., Yang A., Lough L., Brown B., Bashirullah A., Cardozo T., and Ryoo H.D.. 2020. Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat. Commun. 11:4677. 10.1038/s41467-020-18453-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar, S.S., and Brodsky J.L.. 2008. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, R.L., Figueroa A., Court F.A., Thielen P., Molina C., Wirth C., Caballero B., Kiffin R., Segura-Aguilar J., Cuervo A.M., et al. 2012. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet. 21:2245–2262. 10.1093/hmg/dds040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and Ron D.. 2011. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang, T., Lao U., and Edgar B.A.. 2009. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J. Cell Biol. 186:703–711. 10.1083/jcb.200904090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., and Montell C.. 2007. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454:821–847. 10.1007/s00424-007-0251-1 [DOI] [PubMed] [Google Scholar]
- Wang, T., Wang X., Xie Q., and Montell C.. 2008. The SOCS box protein STOPS is required for phototransduction through its effects on phospholipase C. Neuron. 57:56–68. 10.1016/j.neuron.2007.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Punzo C., Ash J.D., and Lobanova E.S.. 2022. Tsc2 knockout counteracts ubiquitin-proteasome system insufficiency and delays photoreceptor loss in retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 119:e2118479119. 10.1073/pnas.2118479119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Chiang W.C., Sumpter R. Jr, Mishra P., and Levine B.. 2017. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 168:224–238.e10. 10.1016/j.cell.2016.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Zhang L., Yang Y., Li N., Lai W., Wang F., Zhu X., and Wang T.. 2020. ER complex proteins are required for rhodopsin biosynthesis and photoreceptor survival in Drosophila and mice. Cell Death Differ. 27:646–661. 10.1038/s41418-019-0378-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., et al. 2015. Histamine Recycling Is Mediated by CarT, a Carcinine Transporter in Drosophila Photoreceptors. PLoS Genetics. 11. 10.1371/journal.pgen.1005764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., and Wang T.. 2016. CULD is required for rhodopsin and TRPL channel endocytic trafficking and survival of photoreceptor cells. J. Cell Sci. 129:394–405. 10.1242/jcs.178764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Zhao H., and Wang T.. 2020. Suppression of retinal degeneration by two novel ERAD ubiquitin E3 ligases SORDD1/2 in Drosophila. PLoS Genet. 16:e1009172. 10.1371/journal.pgen.1009172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C., Liu J., Gao J., Sun Y., Zhang L., Song H., Xue L., Zhan L., Gao G., Ke Z., et al. 2019. IRE1 promotes neurodegeneration through autophagy-dependent neuron death in the Drosophila model of Parkinson’s disease. Cell Death Dis. 10:800. 10.1038/s41419-019-2039-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J., Qiu Y., Frontera E., Jia L., Khan N.W., Klionsky D.J., Ferguson T.A., Thompson D.A., and Zacks D.N.. 2018. Inhibiting autophagy reduces retinal degeneration caused by protein misfolding. Autophagy. 14:1226–1238. 10.1080/15548627.2018.1463121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu, T., Nair U., Yang Z., and Klionsky D.J.. 2006. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281:30299–30304. 10.1074/jbc.M607007200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Matsui T., Yamamoto A., Okada T., and Mori K.. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhang, W., Hietakangas V., Wee S., Lim S.C., Gunaratne J., and Cohen S.M.. 2013. ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes Dev. 27:441–449. 10.1101/gad.201731.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Wang J., and Wang T.. 2018. The V-ATPase V1 subunit A1 is required for rhodopsin anterograde trafficking in Drosophila. Mol. Biol. Cell. 29:1640–1651. 10.1091/mbc.E17-09-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., and Wang T.. 2020. PE homeostasis rebalanced through mitochondria-ER lipid exchange prevents retinal degeneration in Drosophila. PLoS Genet. 16:e1009070. 10.1371/journal.pgen.1009070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Palam L.R., Jiang L., Narasimhan J., Staschke K.A., and Wek R.C.. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283:7064–7073. 10.1074/jbc.M708530200 [DOI] [PubMed] [Google Scholar]
- Zhu, H., Bhatt B., Sivaprakasam S., Cai Y., Liu S., Kodeboyina S.K., Patel N., Savage N.M., Sharma A., Kaufman R.J., et al. 2019. Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 10:1084. 10.1038/s41467-019-08908-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, C.S., Cowman A.F., and Rubin G.M.. 1985. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 40:851–858. 10.1016/0092-8674(85)90344-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the gene expression profiling of retina of the Rh1P37H-GFP/perkRNAi and Rh1P37H-GFP flies related to Fig. 4 A.
shows the gene expression profiling of retina dissected from the ninaE-GFP and ninaE-xbp1-RE flies related to Fig. S3 A.
shows comparison of retinal protein levels of Rh1P37H-GFP perkRNAi with Rh1P37H-GFP flies related to Fig. 5 A.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S2.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
All data are available in the main text or the supplementary materials.