Summary
Background
Long-COVID (LC) encompasses diverse symptoms lasting months after the initial SARS-CoV-2 infection. Symptoms can be debilitating and affect the quality of life of individuals with LC and their families. Although the symptoms of LC are well described, the aetiology of LC remains unclear, and consequently, patients may be underdiagnosed. Identification of LC specific biomarkers is therefore paramount for the diagnosis and clinical management of the syndrome. This scoping review describes the molecular and cellular biomarkers that have been identified to date with potential use for diagnosis or prediction of LC.
Methods
This review was conducted using the Joanna Briggs Institute (JBI) Methodology for Scoping Reviews. A search was executed in the MEDLINE and EMBASE databases, as well as in the grey literature for original studies, published until October 5th, 2022, reporting biomarkers identified in participants with LC symptoms (from all ages, ethnicities, and sex), with a previous infection of SARS-CoV-2. Non-English studies, cross-sectional studies, studies without a control group, and pre-prints were excluded. Two reviewers independently evaluated the studies, extracted population data and associated biomarkers.
Findings
23 cohort studies were identified, involving 2163 LC patients [median age 51.8 years, predominantly female sex (61.10%), white (75%), and non-vaccinated (99%)]. A total of 239 candidate biomarkers were identified, consisting mainly of immune cells, immunoglobulins, cytokines, and other plasma proteins. 19 of the 239 candidate biomarkers identified were evaluated by the authors, by means of receiver operating characteristic (ROC) curves.
Interpretation
Diverse cellular and molecular biomarkers for LC have been proposed. Validation of candidate biomarkers in independent samples should be prioritized. Modest reported performance (particularly in larger studies) suggests LC may encompass many distinct aetiologies, which should be explored e.g., by stratifying by symptom clusters and/or sex.
Funding
Dr. Tebbutt has received funding from the Canadian Institutes of Health Research (177747) to conduct this work. The funding source was not involved in this scoping review, or in the decision to submit this manuscript for publication.
Keywords: Long-COVID (LC), Post-acute sequelae of SARS-CoV-2 infection (PASC), Post-COVID syndrome (PCS), Biomarkers
Research in context.
Evidence before this study
Between 5 and 50% of COVID-19 survivors develop long-COVID (LC; based on global estimates), reducing their quality of life. LC is often underdiagnosed, due to the variable and diverse symptoms experienced. Hence, biomarkers are needed to improve LC patients’ diagnosis and care. Potential biomarkers of LC have been reported in individual studies; however, a scoping review of them has not been conducted. We executed a search strategy for: “long covid”, “post covid”, “biomarker”, “predictor”, and similar terms in MEDLINE, EMBASE, and grey literature databases. We included original cohort studies identifying candidate biomarkers in patients with a diagnosis of LC from any age and ethnicity, published until October 5th, 2022. We excluded studies without a control group, pre-prints, conference proceedings, case reports/series, and non-English studies. Two independent reviewers evaluated the studies and extracted the data.
Added value of this study
To our knowledge, this is the first scoping review to compile the results of studies that have identified molecules or cells in the blood of LC patients that could be potential candidate biomarkers of LC. Our review included 23 studies, which identified 239 candidate biomarkers, and grouped them into molecular types, discussing their biological rationale. In addition, we identified biomarkers for specific symptoms of LC, and synthesized biomarkers that exhibited good performance characteristics, and those that were independently validated. This review paves the way for LC biomarker development, by providing a key database of relevant molecules and cells that should be further studied and validated.
Implications of all the available evidence
This review presents the current landscape of cellular and molecular features associated with LC that could be used as disease biomarkers. Moreover, these candidate biomarkers provide insights into the molecular aetiology of LC, which is currently very limited. The review highlights the research gaps and difficulties, including the need for LC definition harmonization in the research setting, sex and/or ethnicity stratification, as well as symptom-specific and untargeted approaches. The ultimate aim of this review is to facilitate clinical translation and application of biomarkers to improve diagnosis and treatment of this debilitating condition.
Introduction
As of November 2022, the COVID-19 pandemic accounted for more than 636 million confirmed cases and over 6.6 million deaths worldwide.1,2 Vaccination efforts have reduced the severity and mortality of the disease3,4; however, a global estimated range of 5–50% of SARS-CoV-2 infection survivors continue to experience long-term symptoms,5, 6, 7, 8 reducing their quality of life.9 These symptoms are commonly referred to as long-COVID (LC), Post-Acute Sequelae of SARS-CoV-2 Infection (PASC), or Post- COVID Syndrome (PCS). More than 200 diverse symptoms have been reported across diverse systems,10 including the respiratory, neurological, cardiovascular, and gastrointestinal systems.11 In the absence of a universal definition, LC has been defined by the World Health Organization (WHO) as diverse symptoms that are present 3 months after initial infection with SARS-CoV-2, which last for at least 2 months, and which cannot be explained by an alternative diagnosis.12
Systematic reviews and meta-analyses aiming to characterize this condition have identified the most common symptoms of LC as fatigue,11,13, 14, 15, 16, 17, 18, 19, 20, 21, 22 dyspnea,13,15,17, 18, 19, 20, 21, 22 cough,15,17,18,20,22 arthralgia,15,16,20,22 myalgia,20,21 concentration impairment,14,19,21 weakness,14,16,19 effort intolerance,21 breathlessness,14 thoracic pain,15 chest pain,17,18,20 reduced quality of life,11,14,18 depression,19 anxiety,19 sleep disorders,20,21 palpitations,20 anosmia,20,22 ageusia,20,22 and skin problems.11,20 Due to the multiple symptoms, a categorization of LC would allow a better understanding of this ailment. Some studies have categorized LC symptoms into pulmonary/respiratory11,14,16,20 gastrointestinal,11,14,16,20 musculoskeletal,14,16,20 neurological,14,16,20 cardiovascular,11,20 and mental health related.11,14,16 The risk factors for developing LC are primarily female sex,15,17,19,23,24 older age,15,17 comorbidities,15,17 and severity of COVID-19 infection.15,17,23,24 Nonetheless, LC has also been identified in children, young patients, and people who had mild COVID-19.24,25
The complexity of LC and the insufficient knowledge about its aetiology hinder the diagnosis and treatment of these individuals. Biomarkers can play a fundamental role in improving the diagnosis, prognosis, and treatment stratification of people living with LC,11,26,27 and may even suggest novel treatments.
To date, no scoping review of LC biomarkers has been conducted. Molecular and cellular biomarkers of LC have been reported, including immune cells, antibodies, inflammation proteins, endothelium damage markers, and clotting factors.28, 29, 30, 31, 32, 33, 34, 35 Scoping reviews identify and systematically map the available evidence on a given topic; they are useful for examining emerging evidence when it is still unclear what other, more specific questions can be addressed by a systematic review.36 Considering that LC biomarkers are an emerging topic reported by studies with heterogeneous designs and diverse definitions of LC, we conducted a scoping review aiming to compile all the evidence regarding candidate biomarkers of LC with a probable diagnostic/prognostic value for future studies.
Methods
This review was conducted using the updated methodological guidance developed by the Joanna Briggs Institute,37 and reported in the Preferred Reporting Items for Systematic Reviews and Meta-analyses, extension for scoping reviews (PRISMA-ScR) framework.38 The protocol for this review was registered on June 6th, 2022, and is available in the Open Science Framework (https://osf.io/hwfvc).
Eligibility criteria
We included prospective and retrospective cohort studies reporting candidate biomarkers identified in LC patients, with confirmed previous infection of any variant of SARS-CoV-2. Case reports/series, descriptive cross-sectional studies, or studies without a control group were excluded, in order to include only biomarkers that were statistically significant to differentiate LC individuals from control subjects. The timeframe or LC definition was not specified. Pre-prints, non-English studies, book chapters, conference proceedings, editorials/letters, and animal studies were also excluded.
Search strategy
A search strategy was built for MEDLINE (Appendix I), and all identified keywords and index terms were adapted for each included database and/or information source. The search was executed on May 3rd, 2022 (last updated October 5th, 2022) in the MEDLINE and EMBASE databases and complemented with a grey literature search in Grey Matters, des Libris, Trip Database, Google Scholar, the Web of Science, Open Science Frame (OSF), Cochrane Central Register of Controlled Trials (CENTRAL), ClinicalTrials, and International Clinical Trials Registry Platform (ICTRP).
Selection of sources of evidence
The citations identified in the search were uploaded into Covidence, a screening and data extraction tool for undertaking systematic or scoping reviews. Two independent reviewers (E.E. and C.Y.) screened titles and abstracts, as well as the full texts of selected citations based on the inclusion criteria. Reasons for the exclusion of sources of evidence at full text screening were recorded. Disagreements between the reviewers were resolved through discussion and consensus. The search results and the study inclusion process are presented in a PRISMA-ScR flow diagram37 (Fig. 1).
Fig. 1.
PRISMA flow diagram.
Data extraction and synthesis of results
Data from the included studies were extracted by the two independent reviewers using an extraction chart developed by them and added to Covidence. The following items were extracted: first author, country of the study, date of publication, study design, sample size, months from acute infection, LC definition, hospitalization of patients, ethnicity, sex, comorbidities, vaccination status, LC symptoms, molecular or clinical biomarkers, sample for biomarker detection. The data extracted were exported from Covidence to an Excel spreadsheet and analysed in R for descriptive statistics.
Sex/gender information were extracted from data reported in the studies included in this review. In these studies, sex stratification of data was not performed, which should be acknowledged as a limitation since LC condition predominantly affects women.
Statistics
A meta-analysis was not the primary aim of this review due to the heterogeneity of LC definitions, control populations, and demographic variables reported by the studies. Variables of the LC population including size (number of LC individuals), age (years), sex, the severity of infection, ethnicity, vaccination status, co-morbidities, and LC symptoms (percentages) reported by the studies were summarized by median calculation in R, and presented with the corresponding interquartile range (IQR) in Table 1. The summarized data is presented in the text as a median percentage accompanied by the number of studies reporting that variable. Co-morbidities and LC symptoms reported by more than two studies were included in Table 1.
Table 1.
LC population demographics and characteristics.
| Variable | Median (IQR) | No. of studies |
|---|---|---|
| Population size (N) | 67.00 (23.50–151.00) | 23 |
| Age (years) | 51.80 (49.00–55.70) | 13 |
| Sex (Female) % | 61.10 (52.31–70.83) | 18 |
| Vaccination % (LC and controls) | 11.60 | 1 |
| Severity of infection | ||
| Mild % | 62.98 (41.39–82.14) | 6 |
| Moderate % | 23.90 (17.65–34.17) | 4 |
| Severe % | 25.80 (4.55–36.36) | 5 |
| Ethnicity | ||
| White % | 75 (57.89–90.30) | 9 |
| Hispanic/Latino % | 27.50 (15.75–29.52) | 7 |
| Black % | 3.62 (2.47–6.19) | 8 |
| Asian % | 4.06 (0–5.89) | 8 |
| Comorbidities | ||
| Diabetes % | 17.60 (11.00–21.40) | 9 |
| Hypertension % | 27.54 (23.50–31.25) | 8 |
| Asthma/COPD % | 16.20 (11.45–18.25) | 6 |
| Autoimmune disease % | 11.25 (8.07–14.71) | 6 |
| Cancer % | 4.45 (3.00–6.57) | 6 |
| Cardiovascular disease % | 4.28 (3.07–11.89) | 6 |
| Obesity % | 26.65 (13.68–42.75) | 4 |
| Lung disease % | 16.80 (14.34–18.95) | 4 |
| LC symptom | ||
| Fatigue % | 43.80 (31.95–58.15) | 15 |
| Dyspnea % | 30.00 (21.40–51.40) | 13 |
| Anosmia/ageusia % | 20.40 (16.10–26.65) | 12 |
| Cough % | 17.80 (9.00–20.70) | 11 |
| Chest pain % | 17.90 (10.40–25.82) | 10 |
| Myalgia % | 30.80 (24.68–50.40) | 8 |
| Sleep disorder % | 29.05 (11.88–36.45) | 8 |
| Palpitations % | 18.18 (10.35–20.25) | 7 |
| Gastrointestinal symptoms % | 9.00 (6.75–15.00) | 7 |
| Arthralgia % | 33.80 (15.43–48.65) | 6 |
| Concentration/ memory impairment % | 36.80 (34.70–46.70) | 5 |
| Nasal congestion % | 11.60 (5.60–19.20) | 5 |
| Diarrhoea % | 9.09 (8.30–12.30) | 5 |
| Sore throat % | 5.80 (5.60–9.60) | 5 |
| Respiratory symptoms % | 30.04 (12.16–45.70) | 4 |
| Anxiety % | 18.20 (13.25–23.62) | 4 |
| Nausea/indigestion % | 11.20 (7.36–16.40) | 4 |
| Skin problems % | 6.65 (4.60–9.65) | 4 |
| Brain fog % | 71.00 (50.50–78.68) | 3 |
| Depression % | 24.90 (16.60–27.45) | 3 |
| Balance disorder % | 17.80 (14.25–19.45) | 3 |
| Appetite loss % | 15.60 (12.75–16.00) | 3 |
| Stomach pain % | 8.20 (6.60–11.05) | 3 |
| Hair loss % | 5.60 (4.65–7.34) | 3 |
| Chills % | 4.55 (3.62–4.77) | 3 |
| Fever % | 2.70 (2.20–5.89) | 3 |
| Vomiting % | 1.40 (1.10–4.85) | 3 |
The most frequent co-morbidities preceding COVID-19 infection and long-COVID symptoms in LC individuals are presented (reported by >2 studies).
Ethics statement
Ethical approval was not required.
Role of funding source
This scoping review was funded by a grant from the Canadian Institutes of Health Research (No. 177747) received by Dr. Tebbutt. The funding agency was not involved in the study design, data collection, data analyses, interpretation, writing of report, or in the decision to submit this manuscript for publication.
Results
Search, selection, and study characteristics
A total of 2757 articles from the MEDLINE and EMBASE databases, and 86 from the grey literature search were imported to Covidence. Duplicated articles were removed, and 1808 articles remained for screening by two independent reviewers. 81 articles were selected for full-text screening; 23 of these met eligibility criteria and were included, as represented in the PRISMA flow diagram (Fig. 1). Included articles were published between 2021 (8/23; 34.8%) and 2022 (15/23; 65.2%); all were cohort studies, conducted predominantly in the United States (10/23; 43.5%), United Kingdom (2/23; 8.7%), and Spain (2/23; 8.7%). Four studies performed proteomics,39, 40, 41, 42 one metabolomics,43 one multi-omics34 analysis, and the rest conducted assays for specific panels of molecules or cells (Supplementary Table S1).
Characteristics of the populations of the studies
The eligible studies involved a total of 8012 subjects, of which 2211 were LC patients; however, two studies by Peluso et al.31,44 were based on the same 121 individuals, a subgroup of the LIINC cohort (NCT04362150), so the unique population involves 7891 subjects and 2163 LC patients. Despite this, the biomarkers reported in each study by Peluso et al. are different, as well as the case definition and comparator groups construction. Not all the included studies reported all the demographic variables considered in this review; therefore, the number of studies reporting each variable is stated. Across the 23 studies, the median number of LC participants was 67, with a median age of 51.8 years, predominantly female (61.10%), white (75%), and non-vaccinated (99%), except for one study, with 11.6% of the total of participants vaccinated.45 The most reported comorbidities (presented as median percentage across the studies/number of studies reporting the variable) are diabetes (17.60%; 9/23), hypertension (27.54%; 8/23), asthma/COPD (6.20%; 6/23), and autoimmune disease (11.25%; 6/23), among others (Table 1). Cortellini et al.46 study consisted of an oncological cohort (Supplementary Table S1).
LC definition and symptoms
The majority of studies used their own LC definition (15/23; 65.2%); the rest adopted definitions set by the Centers for Disease Control and Prevention (CDC),47 National Institute for Health and Care Excellence (NICE),48 or WHO definitions. The studies varied in their definition of LC as occurring between 133,34,44,45,49, 50, 51 and 1252 months after the acute infection (Supplementary Table S1). The instruments used to diagnose LC consisted primarily of patient-reported outcomes (PRO) questionnaires, study-specific questionnaires,33,34,40,50,51,53, 54, 55, 56 a CDC questionnaire,31,43,44 the NICE guidelines,57 or other criteria.29,46,52 The symptoms of LC most reported across the studies (presented as median percentage across the studies/number of studies reporting the variable) were fatigue (43.8%; 15/23), dyspnea (30.0%; 13/23), anosmia/ageusia (20.40%; 12/23), cough (17.8%; 11/23), and chest pain (17.9%; 10/23), myalgia (30.8%; 8/23), sleep disorder (29.1%; 8/23), among others (Table 1).
LC candidate biomarkers
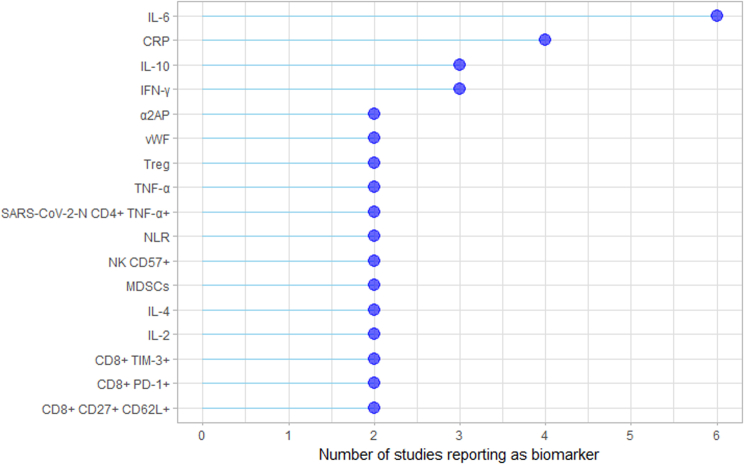
Herein, candidate biomarkers were defined as molecules or cells, for which plasma levels differed significantly between LC patients and the control population (p < 0.05). A total of 239 candidate biomarkers were identified. All of them were detected in blood samples, except for SARS-CoV-2 RNAemia detected in nasal swabs. The most frequently reported biomarkers across the studies were interleukin (IL)-6, C-reactive-protein (CRP), IL-10, interferon (IFN)-γ, α2 antiplasmin (α2AP), von Willebrand factor (vWF), and T regulatory cells (Treg) (Supplementary Fig. S1).
The control groups included subjects recovered (127/239; 53.1%), healthy non-exposed to the virus (90/239; 37.7%), during the acute phase (26/239; 10.9%), and with mild LC (defined by the severity of ongoing physical health, mental health, and cognitive impairment40) (18/239; 7.5%) (Supplementary Table S1).
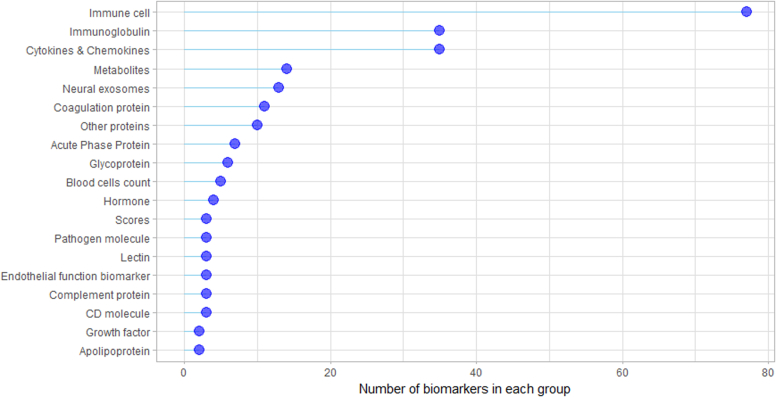
We grouped candidate biomarkers by the type of molecule or cell, in order to facilitate their description. Our groups include immune cells, immunoglobulins, cytokines & chemokines, complement molecules, cluster of differentiation (CD) molecules, lectins, metabolites, neuronal proteins, coagulation proteins, blood cell counts, acute phase proteins (APPs), pathogen molecules, hormones, endothelial function markers, growth factors, other plasma proteins, and LC scores. The groups containing the highest number of biomarkers were immune cells (77/239; 32.2%), immunoglobulins (35/239; 14.6%), and cytokines & chemokines (35/239; 14.6%) (Supplementary Fig. S2).
Validation of LC candidate biomarkers
Biomarker validation is a process used to establish if a biomarker's performance is acceptable for its intended purpose. This process encompasses internal validation (using the data with which a biomarker was developed); external validation (using a completely independent data set); analytical validation (determining sensitivity, specificity, accuracy, and precision); and clinical validation (confirming an association between the biomarker and the end point of interest, and revealing the clinical utility of the biomarker).58
Of the 239 biomarkers identified in this review, only 19 candidate biomarkers (7.9%) were evaluated by receiver operating characteristic (ROC) analysis33,45,46,52 with area under the curve (AUC) values reported. Of these, only a single study externally validated the biomarker consisting of a multi-analyte score,33 and none of them performed clinical validation. Interpretation of AUC is as follows 1.0 is a perfect discriminative biomarker, 0.9–0.99 is an excellent biomarker, 0.8–0.89 is a good biomarker, 0.7–0.79 is a fair biomarker, 0.51–0.69 is a poor biomarker, and ≤0.5 is of no value.59 The candidate biomarkers reported by the studies included in this review oscillate between no discrimination, and excellent discrimination; however, higher AUC values were typically reported in smaller studies (Table 2).
Table 2.
Candidate biomarkers were evaluated by receiver operating characteristic (ROC) analysis, with area under the curve (AUC) values.
| Author/cohort | Candidate biomarker | Performance measured by the study [AUC (95% CI), ACC ±95%CI, or F1 ±95%CI] | Time point |
|---|---|---|---|
| Wu et al. Discovery cohort = 50 (24 Pulmonary LC, 26 Recovered) |
CD8+ CD27+ CD62L+ , NK CD57+ and CD4+ perforin+ | AUC 0.942 | 12 months |
| CD8+ CD27+ CD62L+ | AUC 0.837, SE 0.885, SP 0.708 | ||
| NK CD57+ | AUC 0.819, SE 0.708, SP 0.885 | ||
| CD4+ perforin+ | AUC 0.665, SE 0.875, SP 0.423 | ||
| Cervia et al. Discovery cohort = 134 (85 LC = 85, 49 recovered) Validation cohort = 395 |
LC Score 1 (age, # symptoms acute, history of asthma, IgM, IgG3) |
Discovery cohort AUC 0.771 (0.691–0.851) Validation cohort AUC 0.636 (0.581–0.691) Subgroup hospitalized AUC 0.985 (0.943–1.0) Subgroup outpatients AUC 0.626 (0.569–0.683) |
Acute |
| LC Score 1 (age, # symptoms acute, history of asthma, IgM, IgG3) | AUC 0.743 (0.648–0.838) | 6 months | |
| Cortellini et al. Discovery oncological cohort = 1339 (203 LC, 1136 recovered) |
CRP | AUC 0.66 (0.63–0.69) | Acute |
| NLR | AUC 0.58 (0.55–0.61) | ||
| LDH | AUC 0.57 (0.52–0.61) | ||
| Lionte et al. Discovery cohort = 635 (199 LC, 436 recovered) |
NT-pro BNP | AUC 0.45 (0.36–0.55), SE 0.70, SP 0.80 | Acute |
| CRP | AUC 0.41 (0.36–0.47), SE 0.59, SP 0.71 | ||
| SII | AUC 0.40 (0.35–0.46), SE 0.49, SP 0.64 | ||
| RDW | AUC 0.39 (0.33–0.45), SE 0.55, SP 0.74 | ||
| NLR | AUC 0.38 (0.32–0.43), SE 0.84, SP 0.71 | ||
| Patterson et al. Dataset of LC and recovered individuals split into 60 training/ 20 validation/ 20 test |
LC Score 2 = (IFN-g + IL-2)/CCL4-MIP-1b | F1 0.95, SE 0.975, SP 1.0 | Acute |
| Phetsouphanh et al. Discovery cohort = 62 (31 LC, 31 recovered) |
IFN-β, PTX3 | ACC 0.7854 ±0.0019, F1 0.7736 ±0.0025 | 8 months |
| IFN-β, PTX3, IFN-λ | ACC 0.7968 ±0.0019, F1 0.7852 ±0.0024 | ||
| IFN-β, PTX3, IFN-λ2/3, IL-6 | ACC 0.8159 ±0.0017, F1 0.8053 ±0.0021 | ||
| Panel Cytokines (PD-1, VCAM-1, PECAM-1, ICAM-1, CXCL10, IL-5, sTIM-3, TGF- 1, GM-CSF, MCp-1, IL-12, MPO, IL-13, IL-9, IL-6, ACE-2, IL-10, TNF-α, IL-1β, CXCL9, IL-33, IFN-γ, sCD25, IFN-α2, IFN-λ1, IL-8, IFN-λ2/3, PTX3, IFN-β) | ACC 0.774 ±0.0018, F1 0.7588 ±0.0084 | ||
| Galan et al. Discovery cohort = 50 (30 LC, 20 recovered) |
Random forest algorithm (female, O+, lethargy, pleuritic chest pain, dermatological injuries, T◦, dyspnea, diarrhea, conjunctivitis, autoimmune disease, treated with corticosteroids, antibiotics, and/or vitamin D, total NK cells CD56+, CD3−CD56+CD16+, CD56+NKG2A−NKG2C+, CD56+CD57+NKG2C+; CD3+PD-1+; total CD8+, CD8+ TEMRA, CD8±TCR γδ+; CD4+ Tregs; and cytotoxic activity against NK K562 and/or SARS-CoV-2-Vero E6 cells) | ACC 0.94 ±0.049 | Developed with initial and 11 months data. |
Interpretation of AUC values: AUC = 1.0 perfect, AUC = 0.9–0.99 excellent, AUC = 0.8–0.89 good, AUC = 0.7–0.79 fair, AUC = 0.51–0.69 poor, and AUC ≤ 0.5 of no value. Other biomarkers were evaluated for accuracy (ACC: proportion of participants correctly predicted); and F1 score (a combination of recall [number of LC cases correctly predicted], and precision [proportion of participants predicted to have LC, actually correct]).
SE sensitivity, SP specificity.
Likewise, a LC score based on inflammatory cytokines,60 a random forest algorithm (consisting of demographics, clinical data and immune cell populations),57 and cytokine panels were evaluated for accuracy, defined as the proportion of test participants that had their COVID-19 status correctly predicted; and F1 score, which is a measure that combines recall (how many LC cases were correctly predicted), and precision (of all the participants predicted to have LC, how many were correct)29 (Table 2).
In addition, two studies validated the findings from their primary cohort in an independent cohort, but did not evaluate the performance characteristics of the biomarkers. The Su et al.34 multi-omics study established associations between biomarkers and LC onset in the discovery cohort (209 LC individuals) and validation cohort (100 LC patients) using the same control group in both analysis (457 healthy people). Haffke et al.61 identified endothelial function biomarkers in a discovery cohort of 15 healthy individuals, and 30 LC patients with fatigue symptoms, and confirmed their findings in a second cohort of 70 healthy participants and 56 patients with fatigue LC symptom.
LC candidate biomarkers according to time points of measurement
Samples for biomarker identification were collected at different time points across the studies; the majority (165/239; 69%) were detected in the late convalescent (≥3 months) phase; others (53/239; 22.2%) in the early convalescent (<3 months) phase, and the rest during the acute phase of the infection (36/239; 15.1%) (Fig. 2).
Fig. 2.
Cellular and molecular biomarkers according to time-point measurement. ↑ increased levels of biomarkers in LC population compared to controls. ↓ decreased levels of biomarkers in LC population compared to controls.
Candidate predictive biomarkers for LC at the acute phase
Immune response to viral infection at the acute phase triggers a sustained dysregulation of the immune cells, cytokines, chemokines and immunoglobulins in LC patients. Differential expression of these cells and molecules at primary infection may have a predictive value to identify patients at risk of LC, and add insights for understanding LC molecular pathology.
Regarding cytokines and chemokines, upregulation of IL-2, IL-4, IL-6, IL-10, IL-17, IFN-γ, CCL5/RANTES,60 and CCL360 was detected during the acute phase in individuals who developed LC, compared to control groups. IL-2, which is key in T cell proliferation and self-tolerance,62 and IL-17, which is related to inflammation and autoimmunity, were increased at the acute phase in patients that went on to develop LC, compared to controls.49,60 Similarly, pro-inflammatory IL-662 exhibited higher levels at the acute phase,60 and its higher levels persisted for up to seven months in LC patients from different studies.31,40,42,44,54,60 Elevated levels of IFN-γ, essential for antiviral defence,63 were detected at the initial infection60 and, in another cohort, remained increased in LC individuals for 2 months following infection.31 Regarding IL-4 and IL-10, contradictory findings between some studies were identified. IL-4, considered anti-inflammatory,62 was reduced in patients that developed LC compared to those who recovered fully from COVID-19,49 but increased in another cohort where the control group was healthy controls.60 Likewise, IL-10, which inhibits pro-inflammatory cytokine secretion,62 was increased in patients who developed LC in one cohort,60 but in another study, IL-10 levels were lower in the same group of patients.49 On the other hand, GM-CSF and CCL4 were decreased during COVID-19 infection in patients who later manifested LC, as compared to healthy individuals.60
In regard to immune cells, SARS-CoV-2-specific CD8+ T cells expressing the cytotoxic markers NKG7+ PRF1+ GNLY+ CST7+ were reduced at the acute phase in patients that later displayed gastrointestinal symptoms of LC, but increased in patients that developed respiratory LC symptoms, suggesting different immunopathologies.34 In the above study, activated Tregs at the acute phase, which play a critical role in the maintenance of peripheral tolerance and in controlling the development of autoimmune diseases,64 were enriched in patients developing LC compared to healthy controls.34 In contrast, total Tregs were significantly decreased in individuals who develop LC compared to controls in another study.60 Additionally, increased inflammatory monocytes (CD14+ CD16+ CCR5+)60 and a reduced population of exhausted lymphocytes (CD4+/CD8+ PD-1+) were detected in patients who later suffered LC.60 Interestingly, the inflammatory monocytes remained high during the convalescent period of LC patients, and the exhausted CD4+ and CD8+ lymphocyte populations increased as well.
In addition, immunoglobulins alterations at the acute phase are related to LC onset. Decreased titres of IgM and IgG3 were detected,33 and in a multi-omics study, higher levels of auto-antibodies were associated with the subsequent development of specific LC symptoms (autoAb U1-snRNP with sputum; and autoAb La/SS-B with gastrointestinal symptoms).34
Inflammatory markers, such as C-Reactive Protein (CRP), neutrophil to lymphocyte ratio (NLR), and lactate dehydrogenase (LDH) are routinely measured during COVID-19 infection as severity predictors65; therefore, their utility as LC predictive biomarkers was evaluated. CRP is persistently elevated in LC patients from the acute phase45,46 to seven months after.41,54 An association of increased CRP at the acute phase with a risk of overall LC (OR 2.56 [95% CI 1.67–3.91]) was found in an oncological cohort. However, the authors calculated an unsatisfactory AUC value (0.66 [95% CI 0.63–0.69]) of CRP to differentiate LC patients from the recovered group.46 In the same study higher NLR at the acute phase, was associated with risk of LC onset (OR 1.45 [95% CI 1.01–2.10), but the AUC value (0.58 [95% CI 0.55–0.61]) was poor.46 In another study, lower NLR was detected in individuals who developed LC compared those that recovered from their initial infection, but the AUC calculated was also low (0.38 [95% CI 0.32–0.43]).45 Oncological patients that developed LC exhibited higher levels of LDH at diagnosis, compared to recovered individuals, but its discriminative value for LC in terms of AUC (0.57 [95% CI 0.52–0.61]) was null46 (Table 2). Therefore, this set of inflammatory markers did not demonstrate potential for use as LC predictive biomarkers.
Similar inflammation markers, including red blood cell distribution width (RDW) and systematic inflammation index (SII), calculated as (N × P)/L (where N, P and L represent neutrophil counts, platelet counts and lymphocyte counts, respectively) were evaluated at the acute phase. Higher levels of RDW and SII were identified in patients who developed LC, compared to the recovered group, although low AUC values were determined for both (0.39 [95% CI 0.33–0.45] and 0.40 [95% CI 0.35–0.46], respectively).45 In the same study, reduced levels of N-terminal pro-B-type natriuretic peptide (NT-pro BNP) predicted a higher risk for LC onset (HR 1.68 [95% CI 1.0–2.84]), but the AUC value was not discriminative for LC (0.45 [95% CI 0.36–0.55])45 (Table 2).
Higher levels of the vascular endothelial growth factor (VEGF), which is key in angiogenesis, and was previously identified as a potential biomarker for COVID-19 progression,66 were detected during the acute phase in patients that developed LC.60
SARS-CoV-2 RNAemia at the acute phase was associated with the development of neurologic LC. Likewise, early detection of EBV viremia was associated with subsequent memory impairment, fatigue, and sputum production symptoms of LC in the same study.34
Two scores and one algorithm to predict LC risk at the acute phase have been created. The cytokine based LC Score 2 = (IFN - γ+IL-2)/(CCL4 - MIP-1β) differentiates patients at risk of LC from healthy individuals, and from mild/moderate cases of acute COVID-19 (F1 0.95, Sensitivity 0.975, Specificity 1.0).60 A second LC score based on age, number of symptoms during primary infection, history of bronchial asthma, and an interaction term between IgM and IgG3 levels, is predictive of LC onset at the time of primary infection (AUC 0.77 [95% CI 0.691–0.851]).33 Finally, a random forest algorithm demonstrated an accuracy of 0.94 ± 0.049 to predict LC, based on demographic parameters, clinical factors, and immune response factors (Table 2).57
Candidate biomarkers for LC diagnosis in the convalescent period
The studies included in this review were conducted to decipher the cellular and molecular alterations in patients with LC symptoms compared to controls without LC. The immunophenotypes and molecular signatures of LC participants provide information about LC aetiology and constitute potential biomarkers for LC diagnosis.
Immune cells
Immune cell phenotyping detected an increased frequency of CD8+ TIM-3+ and CD8+ PD-1+ TIM-3+ T cells, in conjunction with increased levels of the soluble TIM-3 marker in LC participants at 3 and 8 months,29 which implies chronic activation of CD8+ T cells considering that PD-1 and TIM-3 are both markers of cells exhaustion. Accordingly, the higher frequencies of CD14+CD16+ inflammatory monocytes, and plasmacytoid dendritic cells (pDCs) expressing the activation markers CD86 and CD38 in these LC patients, suggest a persistent activation of T cells, due to inflammation and/or antigen presentation by activated pDCs or monocytes.29
At 11 months, total CD8+ and its memory subpopulation TEMRA (effector cells essential for CD8+ function) were increased, and naïve CD8+ T cells were reduced in LC patients, suggesting a potent antiviral immune response supported by the increased levels of highly cytotoxic cell populations (CD3+ CD8+ TCR γδ+ cells and CD3+ CD8− TCR γδ+).57 Additionally, the LC group had increased levels of NK cells expressing markers of memory (CD57) and activation (NKG2C), which support a persistent antiviral response.57 In concordance, at 5 months, a sustained response of SARS-CoV-2–S specific CD4+ cell populations expressing the activation marker OX40+ and the exhaustion marker PD-L1+ was found in LC patients, as well as a sustained antigen-specific response of circulating T follicular helper cells (cTfhs)-SARS-CoV-2–S OX40+ PD-L1+.50 Furthermore, the increased levels of Tregs (CD4+ CD25+ CD127low) detected in LC participants 11 months after the acute phase, compared to the recovered group, may point to a failed control of this persistent immune response.57
Cytokines & chemokines
Inflammatory cytokines are upregulated in LC patients. IL-6 exhibited higher levels in overall LC31,42,60 throughout 7 months in different studies, including severe LC patients.40 Likewise, inflammatory IP-10 and TNF-α were increased in LC patients, compared to recovered patients, at 4 months.31 Elevated levels of IFN-γ were detected in the LC group at 2 months.31 In addition, at 11 months higher levels of IFN-β and IFN-λ1 were identified in LC patients.29 Further, the elevation of inflammatory chemokines CXCL1039 and CXCL929 was detected in LC patients60; together, this provides evidence of a chronic inflammatory response in LC. Moreover, the following sets of cytokines 1) IFN-β, PTX3; 2) IFN-β, PTX3, IFN-γ; 3) IFN-β, PTX3, IFN-λ2/3, IL-6; and 4) a 29 cytokines panel were able to differentiate LC patients from asymptomatic controls with accuracy values of 0.7854 ± 0.0019; 0.7968 ± 0.0019; 0.8159 ± 0.0017; and 0.774 ± 0.0018, respectively29 (Table 2).
Only one inhibitor of pro-inflammatory cytokines IL-10 remained higher in LC patients for up to 2 months.31
The FMS-related receptor tyrosine kinase 3 ligand (FLT3LG), which activates hematopoietic progenitors, was significantly increased in severe LC patients at 5 months.40 In these patients an elevated concentration of erythropoietin (EPO) was also detected,40 which is produced in response to hypoxia, and is essential for erythropoiesis.67
Immunoglobulins
Consistent with viral persistence as a possible cause of LC, SARS-CoV-2 specific immunoglobulins were detected in LC patients, and high SARS-CoV-2-IgG titres were associated with LC risk (OR 2.56 [95% CI 1.48–4.38]).53 A higher SARS-CoV-2-S IgG avidity index was observed in LC patients compared to those recovered at 5 months.50 At 12 months titers of SARS-CoV-2 S1/S2 IgG decreased in LC patients; however, increased autoimmunity related immunoglobulins, such as antinuclear antibodies (ANA) were associated with LC (HR 3.37 [95% CI 0.84–13.57]).55 In addition, a proteomic analysis of microclots from blood samples of LC patients identified 23 significantly increased immunoglobulins (Supplementary Table S1).42
Complement proteins
The complement system may be involved in the vascular sequelae of LC, as increased concentrations of the complement components C7 and C6, and the complement factor 1 were detected in plasma microclots of LC patients at 3 months.41
CD molecules
Increased CD83, CD87 (PLAUR), and CD70 were found in severe LC patients.40 CD83 activates macrophages and dendritic cells (DCs), and CD70 is implicated in mediating inflammation of the central nervous system (CNS).40 CD87 is part of the plasminogen activation system, and may be involved in the coagulopathy of LC.68
Lectins
Augmented levels of trefoil factor 2 (TFF2), galectin-9 (LGALS9), and C-type lectin domain family 4 member (CLEC4D) were detected in severe LC patients.40 TFF2 participates in damaged epithelium repair; galectin-9 activates T-cell macrophages and DC,40 and CLEC4D acts as a pattern recognition receptor (PRR).69
Metabolites
A metabolic analysis identified agonists of the N-methyl-d-aspartate (NMDA) receptors with established neurotoxic properties. The kynurenine-to-tryptophan (K/T) ratio and the quinolinic acid–to-tryptophan (Q/T) ratio were higher in LC patients. Elevated levels of S-sulfocysteine were detected in LC patients, and10 other metabolites were also differentially expressed in these patients (Supplementary Table S1).43
Coagulation proteins
Large amyloid deposits (microclots) were detected in plasma samples of LC patients, containing increased levels of coagulation proteins: plasminogen, fibrinogen α, fibrinogen β, α2-antiplasmin (α2AP), von Willebrand factor (vWF), coagulation factors XIII,41 CXCL4, vWF, and α2AP42; and decreased levels of plasma kallikrein (KLKB1).42 These molecules are involved in blood coagulation and fibrinolysis. vWF participates in endothelial injury, hypercoagulation, and may form complexes with CXCL4 released from platelets. α2AP inhibits plasmin, an effector protease of the fibrinolytic system, and KLKB1 digests plasminogen to plasmin.42 These findings, together with the increased levels of inflammatory molecules, highlight the role of coagulation/fibrinolysis in the pathophysiology of LC.
Acute phase proteins (APPs)
APPs respond to inflammatory cytokines. CRP is persistently elevated in LC patients up to 7 months.41,54 Likewise, increased levels of serum Amyloid 1 (SAA1) and SAA4 were identified in the plasma microclots of LC patients at 3 months.41 On the other hand, decreased levels of α-1-acid glycoprotein 1, an inflammation regulator, were found in the microclots of LC individuals at 7 months.42 SAA1 and SAA4 concentrations rise in response to inflammation and tissue injury.70
Pathogen molecules
Increased levels of β-glucan, a marker of fungal plasma translocation, were detected in LC patients.43 LC patients exhibited increased levels of zonulin; although zonulin is not a pathogen molecule, higher levels of this molecule are a marker of fungal and bacterial plasma translocation.43
Hormones
Lower levels of adiponectin were detected in digested microclots of LC individuals compared to the platelet poor plasma of the control group.42 Levels of transforming growth factor (TGF)-α, which induces epithelial development,66 were increased in LC patients.40
Other proteins
Lysosome-associated membrane glycoprotein 3 (LAMP3), expressed in DC,40 was increased in severe LC patients.40 In these patients, follistatin (FST), an inhibitor of the follicle stimulating hormone (FSH),71 was also increased.40 Likewise, the lung surfactant protein72 secretoglobin family 3A member 2 (SCGB3A2) was elevated in severe LC patients.40 Finally, an additional group of 9 diverse proteins was identified in LC patient microclots (Supplementary Table S1).42
Scores
The previously mentioned LC score based on age, symptoms and IgM/IgG3 levels, was found to identify LC patients at 6 months (AUC 0.74 [95% CI 0.648–0.838])33 (Table 2).
Candidate biomarkers for specific LC symptoms
LC is a complex multi-organ condition with different symptoms; therefore, a categorization of this ailment would aid the diagnosis and treatment of patients. While most of the biomarkers identified in this review are intended for overall LC, specific candidate biomarkers for distinct symptoms of LC are grouped in this review into the following categories: pulmonary/respiratory LC (64/239; 26.8%), neurologic LC (22/239; 9.2%), gastrointestinal (GI) LC (8/239; 3.3%), and fatigue LC (4/239; 1.7%) (Fig. 3, Supplementary Table S1).
Fig. 3.
Candidate biomarkers for symptom-specific categories of LC were measured in blood samples, except for SARS-CoV-2 RNAemia, detected in a nasal swab. Abbreviations: MDSCs (myeloid derived suppressor cells), APP (acute phase proteins), CRP (C-reactive protein), WBC (white blood cell counts), MCP-1 (Monocyte chemoattractant protein-1), IL (interleukin), TNF (tumor necrosis factor), IFN (interferon), NK (natural killer), Ig (Immunoglobulin), autoAb (auto-antibody) EBV (Epstein–Barr virus), CMV (cytomegalovirus), N (nucleocapsid SARS-CoV-2 protein), S (spike SARS-CoV-2 protein), GZMB (granzyme B), NDEV (neuron-derived extracellular vesicles), GFAP (glial fibrillary acidic protein), SNPH (syntaphilin), MOTS-c (Mitochondrial open reading frame of the 12S rRNA-c), VDAC1 (voltage-dependent anion-selective channel protein 1), NMDAR1 (N-methyl-d-aspartate receptor 1), MCU (mitochondrial calcium uniporter), NCLX (sodium/calcium exchanger), LETM1 (leucine zipper EF-hand containing transmembrane 1 protein).
Candidate biomarkers for pulmonary and respiratory symptoms of LC
Pulmonary lesions are one of the most common sequelae of COVID-19 infection in the respiratory system. Wu et al. studied a cohort of LC patients with pulmonary sequelae defined as the presence of residual lung lesions at 12 months. The authors delineated a comprehensive profile of immune cells profile (Supplementary Table S1). In summary, they identified enriched populations of CD4+ and CD8+ lymphocytes expressing markers of exhaustion (TIM-3), senescence (CD57) and effector functions (KLRG-1), pointing to chronic excessive inflammation, which leads to simultaneous activation of cytotoxic cell types, including γδT and NK cells with upregulation of degranulation capacity (GZMB+ and perforin+).52 This over-activation of cytotoxic and effector functions may induce lung tissue damage.
Importantly, some of these cell types were evaluated as biomarkers for pulmonary LC. Higher levels of CD8+ CD27− CD62L− cells, a short lived effector subset that results from continuous exposition to viral antigens, were detected in pulmonary LC patients, and correlated with this condition.52 On the other hand, pulmonary LC patients exhibited reduced levels of CD8+ CD27+ CD62L+ T naïve, central memory cells, suggesting an accelerated conversion from naïve to effector lymphocytes in LC syndrome. This cells subset was evaluated as an independent predictor of residual lung lesions (OR 0.738 [95% CI 0.590–0.924]), and a candidate biomarker for pulmonary LC (AUC 0.837).52 In addition, the increased levels of NK cells CD57+ in pulmonary LC patients predicted residual lung lesions (OR 1.181 [95% CI 1.038–1.343]), and had an AUC value of 0.819. Moreover, CD4+perforin+ T cells exhibited a poor discriminative AUC value (0.665), but the combination of the three cell types previously mentioned (CD8+CD27+CD62L+ T cells; CD57+NK cells; and CD4+ perforin+ T cells) improved the AUC value (0.942) for discriminating between pulmonary LC patients and recovered patients52 (Table 2). Additionally, monocytic myeloid-derived suppressor cells (M-MDSCs) percentages correlated with pulmonary LC. Regarding cytokine expression, downregulated IL-2 and upregulated IFN-γ expression were detected in CD8+ of pulmonary LC patients, and CD8+IFN-γ+ percentages correlated with pulmonary LC. The CD4+ T cells of these patients overexpressed IL-17A and IFN-γ, and CD4+IFN-γ+ percentages correlated with pulmonary LC. Furthermore, increased white blood cell counts (WBC), lymphocyte counts, and haemoglobin levels were increased in these patients, the latter suggesting a compensatory mechanism to improve diffusion capacity.52
Another study in pulmonary LC individuals reported higher levels of CRP, IL-6 and SARS-CoV-2-CD4+/CD8+ T cells producing IFN-γ or TNF-α at 7 months.54 Increased CRP at the acute phase was associated with risk of respiratory LC (OR 3.03 [95% CI 1.71–5.36]), in an oncological cohort.46
At 3 months, Su et al.34 determined associations of higher levels of memory-like NK cells with cough symptom of respiratory LC, as well as increased levels of MDSCs with sputum symptom of respiratory LC. They also identified increased autoAb U1-snRNP in patients with sputum, and association of autoAb IFN-α2 and autoAb IFN-α3 with cough and sputum symptoms of respiratory LC.34 Additionally, SARS-CoV-2-specific CD8+ T cells showed enrichment of undifferentiated markers (LEF1, TCF7) in respiratory LC at this time-point, and lower levels of cortisol and cortisone at the acute phase were associated with respiratory LC.34
A metabolite study detected elevated levels of S-sulfocysteine associated with cough symptom of respiratory LC at 5 months.43
Candidate biomarkers for neurologic symptoms of LC
The level of SARS-CoV-2 and mitochondrial proteins in neuron-derived extracellular vesicles (NDEVs) and astrocyte-derived EVs (ADEVs) were assessed in the plasma of patients with neuropsychiatric symptoms of LC at 2 months. Higher levels of exosomes containing SARS-CoV-2 proteins (NDEVs SARS-CoV-2-S1 [RBD], ADEVs SARS-CoV-2-S1 [RBD], NDEVs SARS-CoV-2-N, and ADEVs SARS-CoV-2-N) were detected in these individuals.51
NDEV with mitochondrial proteins [NADH–ubiquinone oxidoreductase (CI-6), cytochrome b-c1 oxidase (CIII-10), mitochondrial open-reading frame of the 12S rRNA-c (MOTS-c), humanin, voltage-dependent anion-selective channel protein 1 (VDAC1), N-methyl-d-aspartate receptor 1 (NMDAR1), mitochondrial calcium uniporter (MCU), sodium/calcium exchanger (NCLX), and leucine zipper EF-hand containing transmembrane 1 protein (LETM1)] were decreased in patients with neuropsychiatric symptoms of LC, except for NDEV syntaphilin (SNPH) which was increased.51 These abnormal NDEV-MP levels have been reported in neurodegenerative and mental illnesses with symptoms similar to neurologic LC.51
In addition, increased glial fibrillary acidic protein (GFAP) protein was detected in neurologic LC patients,44 which could reflect astrocyte dysfunction.44 Agrin (AGRN) was also elevated in severe LC patients,40 its higher concentrations are related to breakdown of the neuromuscular junction, suggesting involvement in neurological and fatigue-related LC.40,73
Higher levels of inflammatory cytokines IL-6, MCP-144 and TNF-α31 were observed in patients with symptoms of neurologic LC.44 In addition, increased SARS-CoV-2-N IgG were detected in neurological LC.34
The previously mentioned metabolite study identified elevated levels of β-glucan in patients with vision problems, sleep problems, neuropathy, and pain. A metabolic analysis identified agonists of the N-methyl-d-aspartate (NMDA) receptors with established neurotoxic properties. The K/T ratio was specifically elevated in patients with neurologic LC manifestations, and elevated levels of S-sulfocysteine were associated with neurocognitive symptoms of LC.43
Candidate biomarkers for gastrointestinal (GI) symptoms of LC
An enrichment of cytotoxic TCR clonotypes of CD8+/CD4+ T cells was observed in patients with GI symptoms of LC, as well as increased SARS-CoV-2-specific CD8+ T cells expressing the cytotoxic markers NKG7+ PRF1+ GNLY+ CST7+ at 2–3 months. In the same study, elevated autoAb La/SS-B levels and persistently increased CMV-specific CD8+ T cells were detected in patients with GI LC.34 Moreover, higher levels of β-glucan were also identified in subjects with GI LC.43
Candidate biomarkers for fatigue symptoms of LC
Endothelial damage may be related to defective vascular function in LC patients. LC patients with fatigue and exertion intolerance had increased levels of endothelin-1 (ET-1), decreased levels of angiopoietin-2 (Ang-2), and a larger peripheral endothelial dysfunction (ED), defined by a diminished reactive hyperaemia index (RHI ≤ 1.67).61 ET-1 mediates vasoconstriction, and Ang-2 participates in endothelial homeostasis and angiogenesis.61 Additionally, EBV viremia at the acute phase was associated with fatigue symptoms of LC.34
Discussion
Long COVID is a post-acute infection syndrome that commonly remains undiagnosed due to its complexity.74 Currently, its diagnosis depends on the identification of LC symptoms by medical professionals.75 Additional complexity arises as LC symptoms affect multiple systems concurrently. Further, some patients experience a constant disease course, while others have relapsing and remitting symptoms.75 Therefore, there is an urgent need to identify biomarkers for LC to enable timely and accurate diagnosis, and to identify the different phenotypes of patients with distinct courses of the disease.
Our scoping review identified 239 molecules/cells that were differentially expressed in LC individuals at different time points, compared to control groups (recovered or healthy participants), from 23 cohort studies. The relevancy of these molecules as potential biomarkers of LC should be further studied and validated; however, they are a starting point for guiding related research and elucidating the molecular pathology of LC.
A total of 19 biomarkers (19/239, 7.9%) were internally validated in four of the included studies by receiver operating characteristic (ROC) analysis,33,45,46,52 and in three studies by accuracy and F1 values.29,57,60 The biomarkers with better performance (AUC 0.942) were based on the combination of exhausted and highly cytotoxic lymphocyte populations (CD8+CD27+CD62L+, NKCD57+ and CD4+perforin+), and were able to differentiate LC pulmonary patients from those who had recovered at 12 months. On the other hand, common inflammation markers, such as CRP, NLR, LDH, NT-pro BNP, SII, and RDW had poor or null AUC values ranging from 0.38 (0.32–0.43) to 0.66 (0.63–0.69) to discriminate between overall LC and recovered populations. It suggests that different symptoms of LC may represent distinct aetiologies, demanding symptom-specific approaches for LC biomarker discovery. However, most of the biomarkers reported by the studies as LC symptom-specific were not validated, hindering more precise analysis of this. Furthermore, multi-analyte biomarker panels demonstrated fair performance to differentiate LC patients from recovered individuals, encompassing cytokine sets with accuracy values ranging from 0.7854 ± 0.0019 to 0.7968 ± 0.001929; and multi-variables scores oscillating between AUC 0.771 (0.691–0.851) and ACC 0.94 ± 0.049.33,60 Nonetheless, the evaluation of these biomarkers did not include external validation or large sample sizes. Therefore, LC biomarker research in larger studies, including external, analytical, and clinical validation, is required.
An optimal biomarker should be objectively quantifiable, sensitive and specific, easily adapted into routine clinical practice, and detectable in easily accessible specimens.58 The pool of candidate biomarkers herein reported were detected in blood samples and therefore could be established in quantifiable assays for clinical practice.
LC biomarkers are needed to stratify risk at SARS-CoV-2 infection onset; confirm LC diagnosis; and/or subset patients for specific interventions.58 The candidate biomarkers measured at the acute phase could have a predictive value for LC risk. These biomarkers consisted mainly of the upregulation of inflammatory chemokines and cytokines (IL-2, IL-4, IL-6, IL-10, IL-17, IFN-γ, CCL5/RANTES, and CCL3); the majority of these have previously been reported as predictors of disease severity due to their participation in the cytokine storm of COVID-19 pathogenesis.76 In addition, some of the candidate biomarkers measured at acute phase may have a biological interplay; for example, reduced levels of IgG3 in LC may be a result of increased IL-4 that impedes Ig isotype switching to IgG3, consequently affecting Fc receptor-dependent viral control.33 Furthermore, higher levels of VEGF at the acute phase60 are promising as LC predictive biomarkers, as further studies have proposed angiogenesis as a key pathophysiological mechanism of LC.77 An important candidate biomarker is the increased cell population of inflammatory monocytes (CD14+CD16+CCR5+),60 which was previously reported as increased in number and metabolic activity per cell in severe COVID-19 patients,78 a cohort that went on to develop LC and was studied by Su et al.34
Regarding the molecular pathology of LC, the candidate biomarkers compiled in this review point to an uncontrolled immune response, triggered by the initial viral infection, and characterized by specific immune signatures. This is evidenced by the higher levels of cytotoxic lymphocytes and NK cells, including granzyme and perforin degranulating lymphocytes,29,52,57 which has been detected in other LC studies for up to a year following infection.79 This suggests a sustained activation of the antiviral immune response,80 consistent with exhausted and senescent phenotypes of T cells, which is common during chronic exposure to antigens.57,79 Likewise, similar novel phenotypes of T cells have been reported to be increased during severe COVID-19, and may persist in LC, such as an exhausted phenotype of CD4+ T cells81; functionally impaired NK cells anti-SARS-CoV-2, which contribute to fibrotic lung disease82; and adaptive-like-/FcR low NK cells.83 In addition, LC patients exhibit higher levels of specific immunoglobulins and lymphocytes against SARS-CoV-2 proteins, emphasizing a possible viral persistence, which has been previously reported in LC patients.84 The sustained activation of the immune response as a result of prolonged exposure to viral antigens leads to the development of autoimmune diseases,79 consistent with the auto-antibodies identified by the studies34,55 included in this scoping review. Furthermore, Tregs, which inhibit the overactivation of T cells and maintain the immune system balance in autoimmune and inflammatory diseases,80 are increased in LC patients for up to 11 months compared to recovered individuals,34,64 but decreased when compared to healthy individuals at the acute phase60 (consistent with another study80). These different Treg levels in LC patients could be explained by recent study findings: different Treg subsets exhibit different dynamics depending on the severity of the acute infection and the early (until 3 months) and late (until 6 months) convalescence periods.85 These studies suggest lower suppressive functions of Treg or an increased influx of naïve Treg in convalescent patients. In addition, a novel phenotype of KIR+CD8+ T cells was found in SARS-CoV-2 infected patients as associated with autoimmune-related complications, suggesting a role of them in controlling autoreactive T cells that are activated during the infection,86 and could have an impact in LC auto-immunity disorders. The high levels of Tregs in LC patients are consistent with immunosuppression, which may promote chronic persistence of SARS-CoV-2.85 All these immune abnormalities ultimately lead to systemic organ damage to the lungs, brain, muscles, and even endothelium, giving rise to LC symptoms.79 Some authors have proposed that IL-6-driven inflammation may disrupt Tregs, leading to mitochondrial dysfunction that constrains neuronal energy metabolism, and could be associated with fatigue and sleeping difficulties.87 Although some pathological traits might be common for different symptom types, several studies report clusters of symptoms in LC patients that imply distinct underlying pathophysiologic mechanisms.88,89 Therefore, biomarkers should also be specific for symptom clusters.
Candidate biomarkers of neurological LC include higher levels of exosomes containing SARS-CoV-2, and decreased levels of mitochondrial proteins, which have been previously related to neurodegenerative diseases.51 Increase in K/T ratio, neuronal proteins such as GFAP44 and AGRN40; and inflammatory cytokines IL-6, MCP-1,44 and TNF-α31 detected in LC suggest neuroinflammation and injury to neurons.43,90 It is critical to discover biomarkers for neurologic LC, as this LC subtype has a delayed onset and longer prevalence (up to 2 years)91 with an unknown prognosis, and possible long-term risk for neurodegenerative diseases due to their neuropathological resemblance92
The GI LC candidate biomarkers herein reported (cytotoxic TCR clonotypes of SARS-CoV-2-specific CD8+ T cells, autoAb La/SS-B levels, CMV-specific CD8+ T cells, and β-glucan higher levels), are consistent with previously proposed mechanisms for this condition as aberrant inflammation, autoimmunity, and viral persistance.93 The biomarkers for fatigue symptoms of LC are related to endothelial damage and defective vascular function (ET-1, Ang-2, endothelial dysfunction). Interestingly, EBV viremia at the acute phase was associated with fatigue symptoms, which is similar to ME-CFS, a post-EBV-infection syndrome. These findings highlight an overlap between clinical signs and biomarkers of LC, ME-CFS, and other post-infection syndromes. However, there are also differences regarding the tropism of the triggering pathogen that demand disease-specific biomarker research.74
Our scoping review has several limitations. First, the 23 studies included had heterogeneous definitions of LC. The absence of a consensus case definition for LC led investigators to adopt 1) a broad case definition that might be overly sensitive in some studies, or 2) a highly specific symptom-severity based definition that may misrepresent milder cases in other studies. Most of the findings herein reported should be assessed taking the common methodological limitations of the studies into consideration. The majority of the included studies had small sample sizes and convenience cohorts that depend on participants returning to follow-up visits. This introduces a selection bias, and in the case of the retrospective oncological cohort of Cortellini et al.,46 a selection bias towards patients with data available. Other studies included cohorts discharged from hospital after COVID-19 which represents a hospitalized population bias. Such a cohort is unlikely to be representative of the general population of individuals recovering from COVID-19 or experiencing LC. Another limitation of the studies was the reliance on self-report to ascertain the presence of LC symptoms. The majority of the studies measured a limited set of biomarkers, and may be missing other cells or molecules relevant to LC pathophysiology. The biomarkers reported were measured in peripheral blood and some of them might not reflect completely the tissue molecular environment of conditions as neurologic LC. All of the studies were conducted in the early stages of the COVID-19 pandemic. It is unclear whether these LC biomarkers are diagnostic or predictive of current SARS-CoV-2 variants, such as Omicron and emerging variants, which have an impact on LC risk.45 In addition, COVID-19 vaccination status was unclear in some of the studies included in this review, preventing the establishment of differences between vaccinated and unvaccinated populations.
In conclusion, this scoping review provides a compilation of candidate molecules and cells that can be used in future studies for LC biomarker discovery, therapeutic target identification, and lead to an increased understanding of the molecular pathology of LC. Moreover, this review highlights gaps and shortcomings in this research field. First, the need for a universal LC definition in terms of onset time, and the harmonization of the instruments for its diagnosis (PROs) in the research setting. This was very heterogeneous across the studies, and we were limited to establish more general conclusions about the biomarkers. Second, considering the fluctuating concentrations of LC biomarkers, the study of their trajectories in LC populations over time should be carried out. Third, the relatively modest performance of the biomarkers, reported in most of the larger studies, suggests that different symptoms may represent distinct aetiologies, demanding symptom-specific approaches for LC biomarker discovery. Fourth, the predominance of the disease in females certainly argues for sex-stratification to be more systematically explored, which was almost completely omitted in the studies. In addition, little is known about LC biomarkers in different ethnicities or genetic predispositions. Finally, the fact that most of the candidate biomarkers are immune cells or molecules advocates for more untargeted approaches in order to identify other molecules such as soluble circulating markers, that may address tissue-specific symptoms not necessarily implicating the immune host response. These comprehensive studies will aid in our understanding of the complex pathology of LC.
Contributors
E.E. contributed to study conceptualisation, data curation, formal analysis, investigation, methodology, visualisation, writing of the original draft, and editing. C.Y. contributed to study conceptualisation, data curation, investigation, and writing, review and editing. S.J.T. contributed to the conceptualisation, supervision, funding acquisition, and writing, review and editing. C.P.S. contributed to the conceptualisation, and writing– review & editing. D.H. contributed to writing, review and editing. S.A. contributed to project administration, and writing, review and editing. Two authors E.E. and C.Y. independently reviewed the studies, extracted the data, and verified it. All authors read and approved the final version of this manuscript.
Data sharing statement
Authors confirm that all relevant data are included in the paper and/or supplementary information files. The data collected for this study can be provided upon reasonable request.
Declaration of interests
All authors have completed the ICMJE uniform disclosure forms. C.Y. reports a grant from the Canadian Institutes of Health Research (grant number: 177747) as payment to their institution to support their work in Canada. The remaining authors have no conflicts of interest to declare.
Acknowledgments
We thank Dr. Vivienne Chan and Dr. Katherine Adolphs from the UBC Centre for Heart Lung Innovation for editorial assistance. This work was supported by a Canadian Institutes of Health Research grant (177747).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104552.
Appendix A. Supplementary data
Supplementary Fig. S1.
Supplementary Fig. S2.
References
- 1.Center for Systems Science and Engineering . Johns Hopkins Coronavirus Resource Center; 2022. COVID-19 Map - Johns Hopkins Coronavirus Resource Center.https://coronavirus.jhu.edu/map.html [Google Scholar]
- 2.World Health Organization WHO coronavirus disease (COVID-19) dashboard with vaccination data | WHO coronavirus (COVID-19) dashboard with vaccination data. World Heal Organ. https://covid19.who.int/ Published online 2022:1-5.
- 3.Richterman A., Meyerowitz E.A., Cevik M. Indirect protection by reducing transmission: ending the pandemic with severe acute respiratory syndrome coronavirus 2 vaccination. Open Forum Infect Dis. 2022;9(2) doi: 10.1093/OFID/OFAB259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal U., Bedston S., McCowan C., et al. Severe COVID-19 outcomes after full vaccination of primary schedule and initial boosters: pooled analysis of national prospective cohort studies of 30 million individuals in England, Northern Ireland, Scotland, and Wales. Lancet. 2022;400(10360):1305–1320. doi: 10.1016/S0140-6736(22)01656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C., Haupert S.R., Zimmermann L., Shi X., Fritsche L.G., Mukherjee B. Global prevalence of post COVID-19 condition or long COVID: a meta-analysis and systematic review. J Infect Dis. 2022 doi: 10.1093/INFDIS/JIAC136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlis R.H., Santillana M., Ognyanova K., et al. Prevalence and correlates of long COVID symptoms among US adults. JAMA Netw Open. 2022;5(10) doi: 10.1001/JAMANETWORKOPEN.2022.38804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collaborators GB of DLC. Hanson S.W., Abbafati C., et al. Estimated global proportions of individuals with persistent fatigue, cognitive, and respiratory symptom clusters following symptomatic COVID-19 in 2020 and 2021. JAMA. 2022;328(16):1604–1615. doi: 10.1001/JAMA.2022.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledford H. How common is long COVID? Why studies give different answers. Nature. 2022;606(7916):852–853. doi: 10.1038/D41586-022-01702-2. [DOI] [PubMed] [Google Scholar]
- 9.Malik P., Patel K., Pinto C., et al. Post-acute COVID-19 syndrome (PCS) and health-related quality of life (HRQoL)—a systematic review and meta-analysis. J Med Virol. 2022;94(1):253. doi: 10.1002/JMV.27309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes L.D., Ingram J., Sculthorpe N.F. More than 100 persistent symptoms of SARS-CoV-2 (long COVID): a scoping review. Front Med. 2021;8:2028. doi: 10.3389/FMED.2021.750378/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groff D., Sun A., Ssentongo A.E., et al. Short-term and long-term rates of postacute sequelae of SARS-CoV-2 infection: a systematic review. JAMA Netw Open. 2021;4(10) doi: 10.1001/JAMANETWORKOPEN.2021.28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 [DOI] [PMC free article] [PubMed]
- 13.Lopez-Leon S., Wegman-Ostrosky T., Perelman C., et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci Rep. 2021;11(1):16144. doi: 10.1038/S41598-021-95565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michelen M., Manoharan L., Elkheir N., et al. Characterising long COVID: a living systematic review. BMJ Glob Health. 2021;6(9) doi: 10.1136/BMJGH-2021-005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen N.N., Hoang V.T., Dao T.L., Dudouet P., Eldin C., Gautret P. Clinical patterns of somatic symptoms in patients suffering from post-acute long COVID: a systematic review. Eur J Clin Microbiol Infect Dis. 2022;41(4):515. doi: 10.1007/S10096-022-04417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long Q., Li J., Hu X., Bai Y., Zheng Y., Gao Z. Follow-ups on persistent symptoms and pulmonary function among post-acute COVID-19 patients: a systematic review and meta-analysis. Front Med. 2021;8:702635. doi: 10.3389/FMED.2021.702635/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera Martimbianco A.L., Pacheco R.L., Bagattini Â.M., Riera R. Frequency, signs and symptoms, and criteria adopted for long COVID-19: a systematic review. Int J Clin Pract. 2021;75(10):14357. doi: 10.1111/IJCP.14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Ramirez D.C., Normand K., Yang Z., Torres-Castro R. Long-term impact of COVID-19: a systematic review of the literature and meta-analysis. Biomedicines. 2021;9(8):900. doi: 10.3390/BIOMEDICINES9080900/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Q., Zheng B., Daines L., Sheikh A. Long-term sequelae of COVID-19: a systematic review and meta-analysis of one-year follow-up studies on post-COVID symptoms. Pathogens. 2022;11(2):269. doi: 10.3390/PATHOGENS11020269/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennings G., Monaghan A., Xue F., Mockler D., Romero-Ortuño R. A systematic review of persistent symptoms and residual abnormal functioning following acute covid-19: ongoing symptomatic phase vs. post-covid-19 syndrome. J Clin Med. 2021;10(24):5913. doi: 10.3390/JCM10245913/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkodaymi M.S., Omrani O.A., Fawzy N.A., et al. Prevalence of post-acute COVID-19 syndrome symptoms at different follow-up periods: a systematic review and meta-analysis. Clin Microbiol Infect. 2022;28(5):657. doi: 10.1016/J.CMI.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-de-las-Peñas C., Palacios-Ceña D., Gómez-Mayordomo V., et al. Prevalence of post-COVID-19 symptoms in hospitalized and non-hospitalized COVID-19 survivors: a systematic review and meta-analysis. Eur J Intern Med. 2021;92:55. doi: 10.1016/J.EJIM.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salamanna F., Veronesi F., Martini L., Landini M.P., Fini M. Post-COVID-19 syndrome: the persistent symptoms at the post-viral stage of the disease. A systematic review of the current data. Front Med. 2021;8:653516. doi: 10.3389/FMED.2021.653516/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maglietta G., Diodati F., Puntoni M., et al. Prognostic factors for post-COVID-19 syndrome: a systematic review and meta-analysis. J Clin Med. 2022;11(6):1541. doi: 10.3390/JCM11061541/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Kessel S.A.M., Olde Hartman T.C., Lucassen P.L.B.J., van Jaarsveld C.H.M. Post-acute and long-COVID-19 symptoms in patients with mild diseases: a systematic review. Fam Pract. 2022;39(1):159. doi: 10.1093/FAMPRA/CMAB076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Haupert S.R., Zimmermann L., Shi X., Fritsche L.G., Mukherjee B. Global prevalence of post-acute sequelae of COVID-19 (PASC) or long COVID: a meta-analysis and systematic review. medRxiv. 2021 doi: 10.1101/2021.11.15.21266377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips S., Williams M.A. Confronting our next national health disaster — long-haul Covid. N Engl J Med. 2021;385(7):577–579. doi: 10.1056/NEJMP2109285. [DOI] [PubMed] [Google Scholar]
- 28.Townsend L., Dyer A.H., Naughton A., et al. Longitudinal analysis of COVID-19 patients shows age-associated T cell changes independent of ongoing ill-health. Front Immunol. 2021;12:1593. doi: 10.3389/FIMMU.2021.676932/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phetsouphanh C., Darley D.R., Wilson D.B., et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol. 2022;23(2):210–216. doi: 10.1038/S41590-021-01113-X. [DOI] [PubMed] [Google Scholar]
- 30.Glynne P., Tahmasebi N., Gant V., Gupta R. Long COVID following mild SARS-CoV-2 infection: characteristic T cell alterations and response to antihistamines. J Investig Med. 2022;70(1):61–67. doi: 10.1136/JIM-2021-002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peluso M.J., Lu S., Tang A.F., et al. Markers of immune activation and inflammation in individuals with postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;224(11):1839–1848. doi: 10.1093/INFDIS/JIAB490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazza M.G., Palladini M., De Lorenzo R., et al. Persistent psychopathology and neurocognitive impairment in COVID-19 survivors: effect of inflammatory biomarkers at three-month follow-up. Brain Behav Immun. 2021;94:138–147. doi: 10.1016/J.BBI.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cervia C., Zurbuchen Y., Taeschler P., et al. Immunoglobulin signature predicts risk of post-acute COVID-19 syndrome. Nat Commun. 2022;13(1):446. doi: 10.1038/s41467-021-27797-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Y., Yuan D., Chen D.G., et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881–895.e20. doi: 10.1016/J.CELL.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fogarty H., Townsend L., Morrin H., et al. Persistent endotheliopathy in the pathogenesis of long COVID syndrome. J Thromb Haemost. 2021;19(10):2546. doi: 10.1111/JTH.15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munn Z., Peters M.D.J., Stern C., Tufanaru C., McArthur A., Aromataris E. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med Res Methodol. 2018;18(1):1–7. doi: 10.1186/S12874-018-0611-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters M.D.J., Marnie C., Tricco A.C., et al. Updated methodological guidance for the conduct of scoping reviews. JBI Evid Synth. 2020;18(10):2119–2126. doi: 10.11124/JBIES-20-00167. [DOI] [PubMed] [Google Scholar]
- 38.Tricco A.C., Lillie E., Zarin W., et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann Intern Med. 2018;169(7):467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J., Schank M., Wang L., et al. Plasma biomarkers for systemic inflammation in COVID-19 survivors. Proteomics Clin Appl. 2022;16(5) doi: 10.1002/PRCA.202200031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans R.A., Leavy O.C., Richardson M., et al. Clinical characteristics with inflammation profiling of long COVID and association with 1-year recovery following hospitalisation in the UK: a prospective observational study. Lancet Respir Med. 2022;10(8):761–775. doi: 10.1016/S2213-2600(22)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pretorius E., Vlok M., Venter C., et al. Persistent clotting protein pathology in Long COVID/post-acute sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc Diabetol. 2021;20(1):1–18. doi: 10.1186/S12933-021-01359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruger A., Vlok M., Turner S., et al. Proteomics of fibrin amyloid microclots in long COVID/post-acute sequelae of COVID-19 (PASC) shows many entrapped pro-inflammatory molecules that may also contribute to a failed fibrinolytic system. Cardiovasc Diabetol. 2022;21(1):190. doi: 10.1186/S12933-022-01623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giron L.B., Peluso M.J., Ding J., et al. Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-κB signaling. JCI Insight. 2022;7(15) doi: 10.1172/JCI.INSIGHT.160989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peluso M.J., Sans H.M., Forman C.A., et al. Plasma markers of neurologic injury and inflammation in people with self-reported neurologic postacute sequelae of SARS-CoV-2 infection. Neurol Neuroimmunol Neuroinflamm. 2022;9(5) doi: 10.1212/NXI.0000000000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lionte C., Sorodoc V., Haliga R.E., et al. Inflammatory and cardiac biomarkers in relation with post-acute COVID-19 and mortality: what we know after successive pandemic waves. Diagnostics. 2022;12(6):1373. doi: 10.3390/DIAGNOSTICS12061373/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortellini A., Gennari A., Pommeret F., et al. COVID-19 sequelae and the host proinflammatory response: an analysis from the OnCovid registry. J Natl Cancer Inst. 2022;114(7):979–987. doi: 10.1093/JNCI/DJAC057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention . 2021. Post-COVID conditions | CDC. Sav Lives, Prot People.https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html [Google Scholar]
- 48.(NICE) NI for H and CE . 2021. Overview | COVID-19 rapid guideline: managing the long-term effects of COVID-19 | Guidance | NICE; pp. 1–111. [Google Scholar]
- 49.Queiroz M.A.F., Neves P.F.M.D., Lima S.S., et al. Cytokine profiles associated with acute COVID-19 and Long COVID-19 syndrome. Front Cell Infect Microbiol. 2022;12:1. doi: 10.3389/FCIMB.2022.922422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Files J.K., Sarkar S., Fram T.R., et al. Duration of post–COVID-19 symptoms is associated with sustained SARS-CoV-2–specific immune responses. JCI Insight. 2021;6(15) doi: 10.1172/JCI.INSIGHT.151544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peluso M.J., Deeks S.G., Mustapic M., et al. SARS-CoV-2 and mitochondrial proteins in neural-derived exosomes of COVID-19. Ann Neurol. 2022;91(6):772–781. doi: 10.1002/ANA.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J., Tang L., Ma Y., et al. Immunological profiling of COVID-19 patients with pulmonary sequelae. mBio. 2021;12(5) doi: 10.1128/MBIO.01599-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peghin M., Palese A., Venturini M., et al. Post-COVID-19 symptoms 6 months after acute infection among hospitalized and non-hospitalized patients. Clin Microbiol Infect. 2021;27(10):1507–1513. doi: 10.1016/J.CMI.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Littlefield K.M., Watson R.O., Schneider J.M., et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PLoS Pathog. 2022;18(5) doi: 10.1371/JOURNAL.PPAT.1010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Abellán J., Fernández M., Padilla S., et al. Immunologic phenotype of patients with long-COVID syndrome of 1-year duration. Front Immunol. 2022;13:4801. doi: 10.3389/FIMMU.2022.920627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peluso M.J., Deitchman A.N., Torres L., et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021;36(6):109518. doi: 10.1016/J.CELREP.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galán M., Vigón L., Fuertes D., et al. Persistent overactive cytotoxic immune response in a Spanish cohort of individuals with long-COVID: identification of diagnostic biomarkers. Front Immunol. 2022;13:1. doi: 10.3389/FIMMU.2022.848886/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ou F.S., Michiels S., Shyr Y., Adjei A.A., Oberg A.L. Biomarker discovery and validation: statistical considerations. J Thorac Oncol. 2021;16(4):537–545. doi: 10.1016/J.JTHO.2021.01.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carter J.V., Pan J., Rai S.N., Galandiuk S. ROC-ing along: evaluation and interpretation of receiver operating characteristic curves. Surgery. 2016;159(6):1638–1645. doi: 10.1016/J.SURG.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 60.Patterson B.K., Guevara-Coto J., Yogendra R., et al. Immune-based prediction of COVID-19 severity and chronicity decoded using machine learning. Front Immunol. 2021;12:700782. doi: 10.3389/FIMMU.2021.700782/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haffke M., Freitag H., Rudolf G., et al. Endothelial dysfunction and altered endothelial biomarkers in patients with post-COVID-19 syndrome and chronic fatigue syndrome (ME/CFS) J Transl Med. 2022;20(1):1–11. doi: 10.1186/S12967-022-03346-2/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62. doi: 10.1016/J.CYTOGFR.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gadotti A.C., de Castro Deus M., Telles J.P., et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020;289:198171. doi: 10.1016/J.VIRUSRES.2020.198171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su H., Longhi M.S., Wang P., Vergani D., Ma Y. Human CD4+CD25(high)CD127 (low/neg) regulatory T cells. Methods Mol Biol. 2012;806:287–299. doi: 10.1007/978-1-61779-367-7_20. [DOI] [PubMed] [Google Scholar]
- 65.Szarpak L., Ruetzler K., Safiejko K., et al. Lactate dehydrogenase level as a COVID-19 severity marker. Am J Emerg Med. 2021;45:638. doi: 10.1016/J.AJEM.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong Y., Han J., Wu X., et al. A novel biomarker for detection of COVID-19 progression. Crit Care. 2020;24(1):1–4. doi: 10.1186/S13054-020-03079-Y/FIGURES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grasso G., Sfacteria A., Cerami A., Brines M. Erythropoietin as a tissue-protective cytokine in brain injury: what do we know and where do we go? Neuroscientist. 2004;10(2):93–98. doi: 10.1177/1073858403259187. [DOI] [PubMed] [Google Scholar]
- 68.Alfano D., Franco P., Stoppelli M.P. Modulation of cellular function by the urokinase receptor signalling: a mechanistic view. Front Cell Dev Biol. 2022;10:705. doi: 10.3389/FCELL.2022.818616/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clec4d - C-type lectin domain family 4 member D - Mus musculus (Mouse) | UniProtKB | UniProt. https://www.uniprot.org/uniprotkb/Q9Z2H6/entry
- 70.Jumeau C., Awad F., Assrawi E., et al. Expression of SAA1, SAA2 and SAA4 genes in human primary monocytes and monocyte-derived macrophages. PLoS One. 2019;14(5) doi: 10.1371/JOURNAL.PONE.0217005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.FST - Follistatin - Homo sapiens (human) | UniProtKB | UniProt. https://www.uniprot.org/uniprotkb/P19883/entry
- 72.SCGB3A2 secretoglobin family 3A member 2 [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/gene/117156
- 73.AGRN - Agrin - Homo sapiens (human) | UniProtKB | UniProt. https://www.uniprot.org/uniprotkb/O00468/entry
- 74.Choutka J., Jansari V., Hornig M., Iwasaki A. Unexplained post-acute infection syndromes. Nat Med. 2022;28(5):911–923. doi: 10.1038/s41591-022-01810-6. [DOI] [PubMed] [Google Scholar]
- 75.Greenhalgh T., Sivan M., Delaney B., Evans R., Milne R. Long covid—an update for primary care. BMJ. 2022;378 doi: 10.1136/BMJ-2022-072117. [DOI] [PubMed] [Google Scholar]
- 76.Del Valle D.M., Kim-Schulze S., Huang H.H., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel M.A., Knauer M.J., Nicholson M., et al. Elevated vascular transformation blood biomarkers in long-COVID indicate angiogenesis as a key pathophysiological mechanism. Mol Med. 2022;28(1):122. doi: 10.1186/S10020-022-00548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee J.W., Su Y., Baloni P., et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat Biotechnol. 2022;40(1):110–120. doi: 10.1038/S41587-021-01020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitsuyama Y., Yamakawa K., Kayano K., et al. Residual persistence of cytotoxicity lymphocytes and regulatory T cells in patients with severe coronavirus disease 2019 over a 1-year recovery process. Acute Med Surg. 2022;9(1) doi: 10.1002/AMS2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadeghi A., Tahmasebi S., Mahmood A., et al. Th17 and Treg cells function in SARS-CoV2 patients compared with healthy controls. J Cell Physiol. 2021;236(4):2829–2839. doi: 10.1002/JCP.30047. [DOI] [PubMed] [Google Scholar]
- 81.Su Y., Chen D., Yuan D., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6):1479–1495.e20. doi: 10.1016/J.CELL.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krämer B., Knoll R., Bonaguro L., et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity. 2021;54(11):2650–2669.e14. doi: 10.1016/J.IMMUNI.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shemesh A., Su Y., Calabrese D.R., et al. Diminished cell proliferation promotes natural killer cell adaptive-like phenotype by limiting FcεRIγ expression. J Exp Med. 2022;219(11) doi: 10.1084/JEM.20220551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaebler C., Wang Z., Lorenzi J.C.C., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiech M., Chroscicki P., Swatler J., et al. Remodeling of T cell dynamics during long COVID is dependent on severity of SARS-CoV-2 infection. Front Immunol. 2022;13:2362. doi: 10.3389/FIMMU.2022.886431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J., Zaslavsky M., Su Y., et al. KIR+CD8+ T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science. 2022;376(6590) doi: 10.1126/SCIENCE.ABI9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kappelmann N., Dantzer R., Khandaker G.M. Interleukin-6 as potential mediator of long-term neuropsychiatric symptoms of COVID-19. Psychoneuroendocrinology. 2021;131:105295. doi: 10.1016/J.PSYNEUEN.2021.105295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kenny G., McCann K., O'Brien C., et al. Identification of distinct long COVID clinical phenotypes through cluster analysis of self-reported symptoms. Open Forum Infect Dis. 2022;9(4) doi: 10.1093/OFID/OFAC060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang H., Zang C., Xu Z., et al. Data-driven identification of post-acute SARS-CoV-2 infection subphenotypes. Nat Med. 2022;29(1):226–235. doi: 10.1038/s41591-022-02116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davis H.E., McCorkell L., Vogel J.M., Topol E.J. Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol. 2023;21(3):133–146. doi: 10.1038/s41579-022-00846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taquet M., Sillett R., Zhu L., et al. Neurological and psychiatric risk trajectories after SARS-CoV-2 infection: an analysis of 2-year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatry. 2022;9(10):815–827. doi: 10.1016/S2215-0366(22)00260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reiken S., Sittenfeld L., Dridi H., Liu Y., Liu X., Marks A.R. Alzheimer’s-like signaling in brains of COVID-19 patients. Alzheimers Dement. 2022;18(5):955–965. doi: 10.1002/ALZ.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meringer H., Mehandru S. Gastrointestinal post-acute COVID-19 syndrome. Nat Rev Gastroenterol Hepatol. 2022;19(6):345–346. doi: 10.1038/s41575-022-00611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.