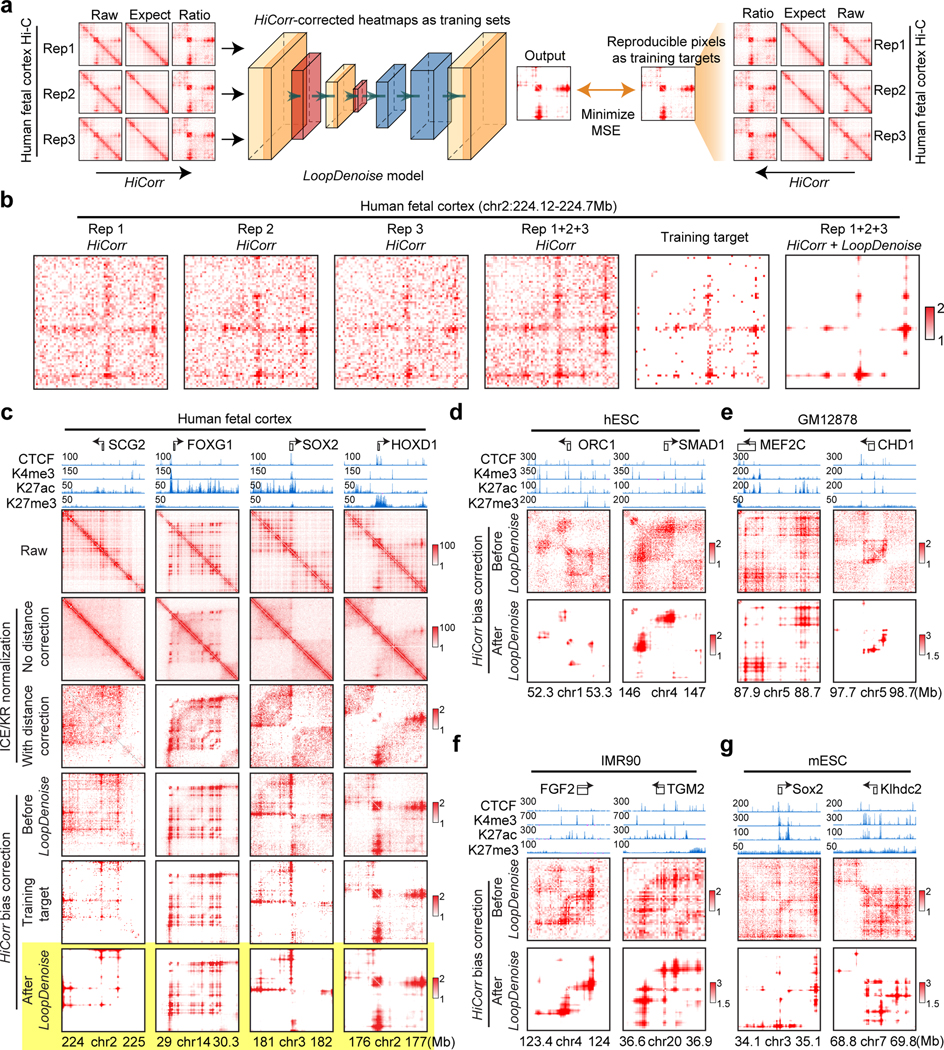
Figure 1. HiCorr and LoopDenoise reveal chromatin loops from noisy Hi-C datasets.
a, Scheme showing the LoopDenoise model architecture and training. The three HiCorr-corrected human fetal brain datasets are used as training sets. The training targets are the reproducible pixels in the heatmaps from the pooled data. b, The example heatmaps from human fetal cortex Hi-C data, including three HiCorr-corrected replicates, pooled data, training target and output from LoopDenoise. c, LoopDenoise performance in the training human fetal cortex Hi-C data at 4 loci. Heatmaps of raw and various processed data are compared. Highlighted row is LoopDenoise output. d-g, Heatmaps showing the application of LoopDenoise to four independent Hi-C datasets in hESCs, GM12878, IMR90, and mESCs. The ChIP-seq tracks show raw reads pileup. See Methods for information how to determine the color scale of each heatmap.