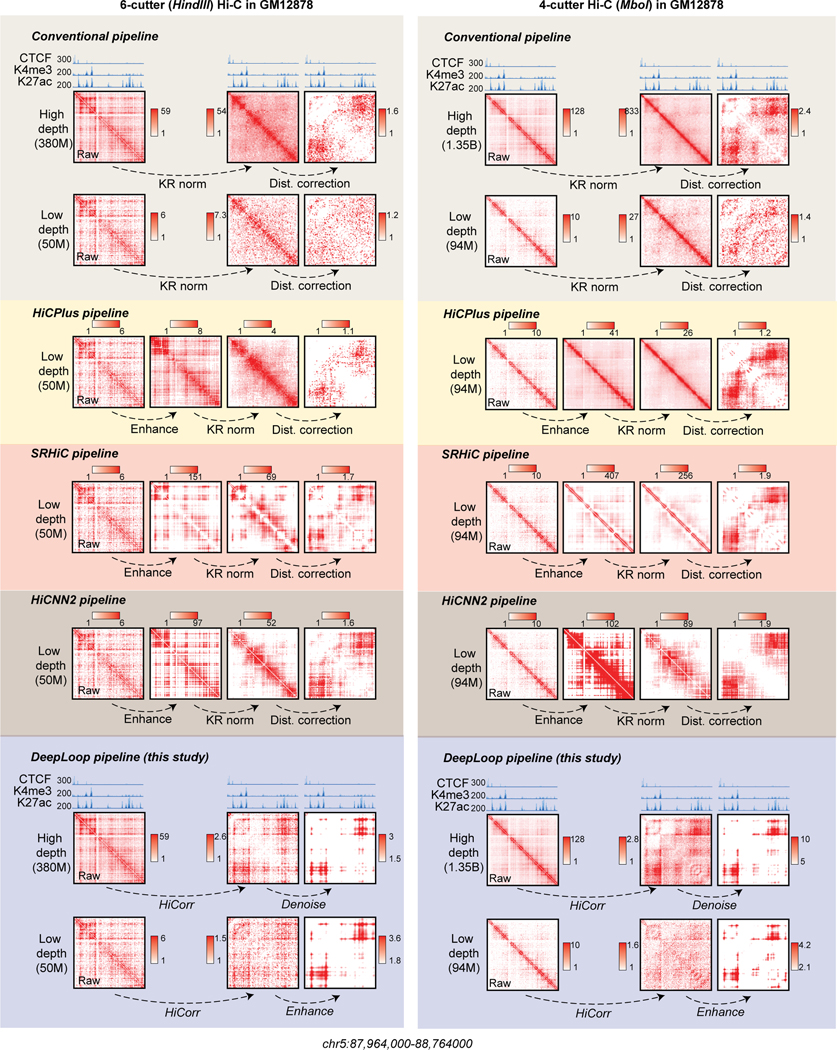
Extended Data Fig. 4. Compare the performance of different pipelines on 6-cutter and 4-cutter Hi-C data in GM12878 cells.
For 4-cutter Hi-C datasets, we chose a 94M down-sampled dataset (1/16 of the original depth) used in HiCPlus, HiCNN2 and SRHiC studies, and the 1.35 billion full-depth as reference. For 6-cutter Hi-C datasets, we chose a 50M down-sampled dataset and the 380M full-depth as reference. For locus chr5:87,964,000–88,764000, the left side showed the contact heatmaps from 6-cutter (HindIII) GM12878 Hi-C processed by different pipelines (colored in background). The right side showed the 4-cutter (MboI) GM12878 Hi-C. The height of browser tracks indicating the raw counts of ChIP-seq.