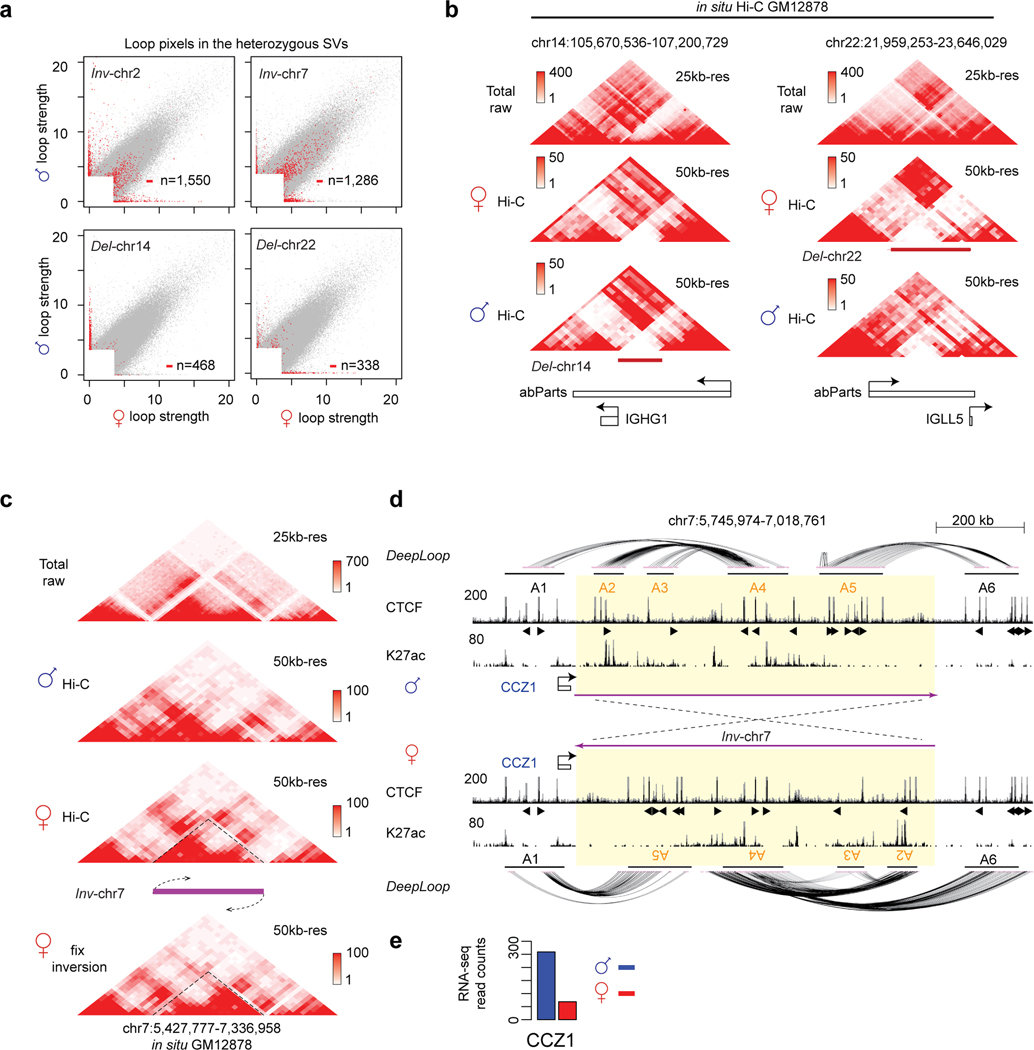
Extended Data Fig. 8. Large heterozygous deletions and inversions detected by allelic DeepLoop analysis.
a, The scatterplots highlight the loop pixels within the entire four SVs region (two inversions and two deletions). b, The contact heatmaps of paternal deletion Del-chr14 and maternal deletion Del-chr22. c, The contact heatmaps of Inv-chr7. d, The genome track of Inv-chr7 shows the chromatin interactions, CTCF and H3K27ac binding on the un-inverted allele and ‘inversion-fix’ allele. In this region, the un-inverted paternal genome has A1-A4 and A5-A6 cross-boundary CTCF loops. The maternal inversion created new A1-A5 and A4-A6 cross-boundary loops due to the inverted orientation the CTCF motifs. Note that in paternal genome, the A1-A4 loop encompass multiple enhancers, while in the inverted maternal genome the A1-A5 loop lack enhancers. e, The gene expression level of gene CCZ1 in two alleles. The height of browser tracks indicating the raw counts of ChIP-seq.