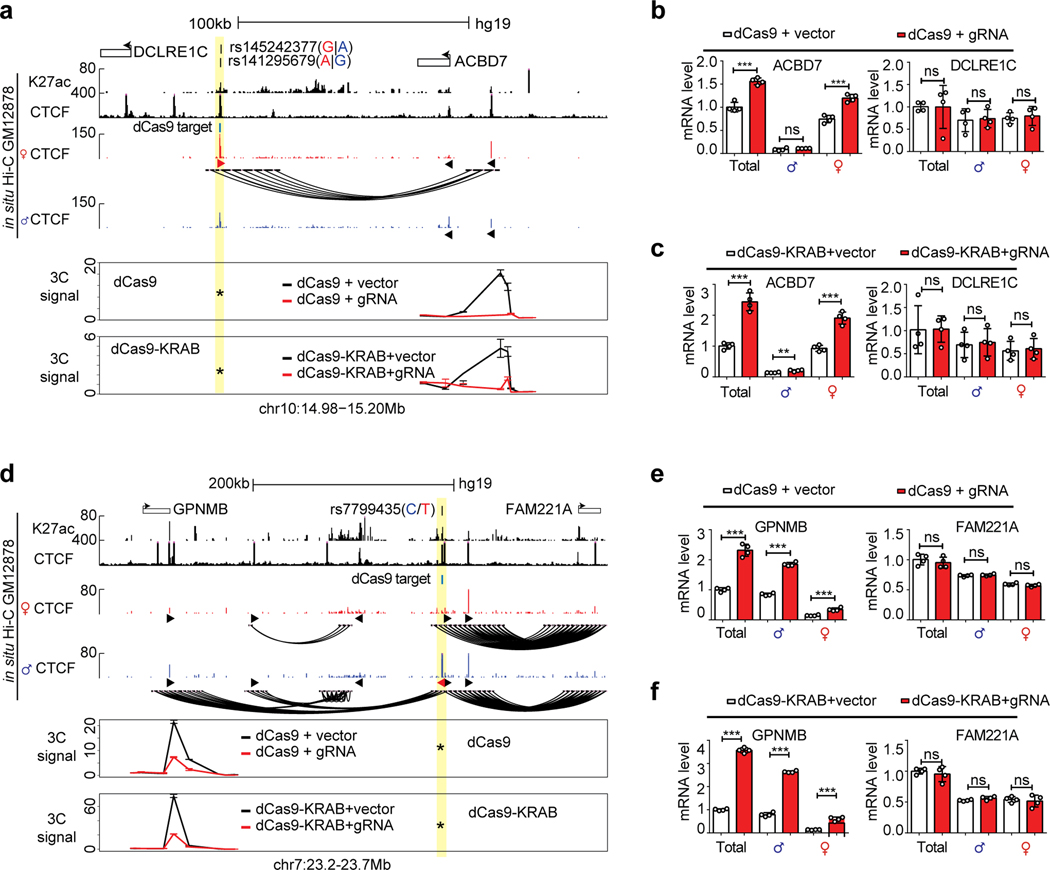
Extended Data Fig. 10. Allele-specific chromatin loops regulate gene expression.
a, 3C assays showing the loss of chromatin loop between the SNP (highlight in yellow) and ACBD7 locus after displacing CTCF binding with either dCas9-KRAB or dCas9 protein. b,c, Bar plots showing the changes of allelic gene expression upon blocking CTCF loops with dCas9 or dCas9-KRAB. d–f, CTCF blocking experiments at GPNMB locus. n = 2 biologically independent experiments. All data are presented as means ± SEM from 4 replicated experiments. **P < 0.01, ***P < 0.001. NS, no significant difference. Two-sided Wilcoxon test. The height of browser tracks indicating the raw counts of ChIP-seq.