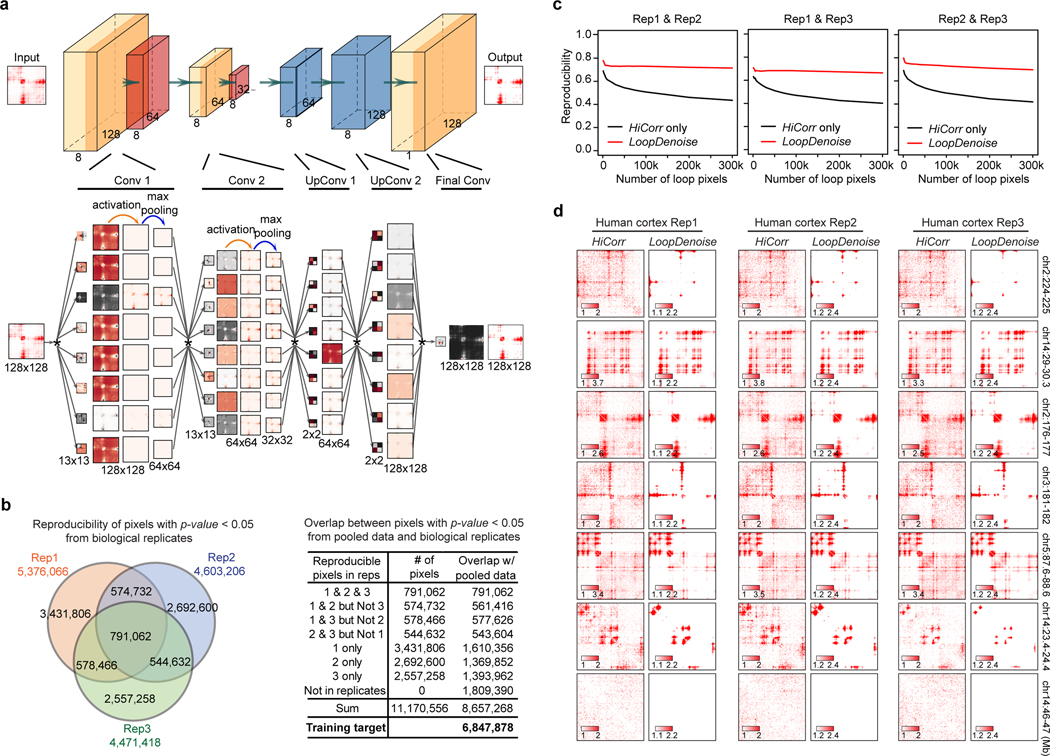
Extended Data Fig. 1. LoopDenoise training procedure, performance and visualization.
a, Detailed LoopDenoise convolutional autoencoder model architecture showing five convolution layers, two in the encoding path using eight 13 × 13 filters, two transpose convolution layers in the decoding path using eight 2 × 2 filters and one final convolution layer using a single 13 × 13 filter. The matrices dimensions of each layer output were also shown. Each layer is visualized by the filters used, the output of convolving the input with this filter, the result of applying ReLU activation and the result of max pooling. The convolution operation is denoted by *. b, Venn diagram showing the reproducible loop pixels between three human fetal brain replicates. The table showing the number of overlapped pixels between significant pixels in the pooled data and each part of pixels shown in the Venn diagram. The pixels that are significant in both pooled data and at least one of the three replicates are the training target in the LoopDenoise model (P < 0.05, negative binomial test). The significance of loop pixels come from the negative binomial test wrapped in HiCorr package. c, Pairwise reproducibility at pixel level (defined as the fraction of common ones when calling the same number of loop pixels from two datasets) between biological replicates of human fetal cortex Hi-C data, when the same numbers of the loop pixels were called. d, The heatmap examples from 7 locus in three human fetal brain replicates, and LoopDenoise output showing more reproducible contact patterns.