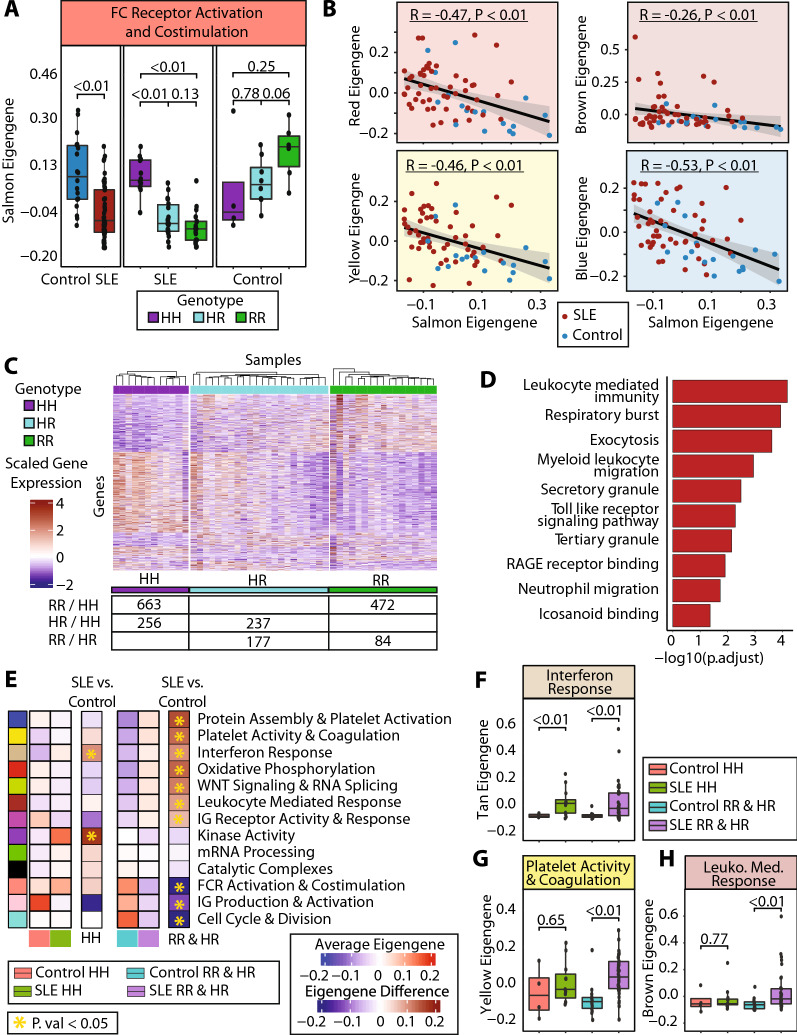
Fig. 4.
The Low-Binding FcγRIIA Recessive Allele Decreases FcγRIIA Signaling Pathways Leading to exacerbated SLE phenotypes A Gene module scores for the Salmon (FC Receptor Activation and Costimulation) for our cohort when divided by SLE vs control, and the SLE when split by genotype and Controls when split by genotype. B Correlation of our Salmon gene module expression vs the Red (Oxidative Phosphorylation), Brown (Leukocyte Mediated Response), Yellow (Platelet Activity and Coagulation), and Blue (Protein Assembly and Localization) gene modules. C Heatmap of SLE samples and genes differentially expressed when comparing HH vs RR, HH vs HR, and HR vs RR. The number of genes differentially expressed in each comparison are shown in the accompanying subtable (RR vs HH, 663 genes up in HH, 472 genes up in RR; HR vs HH, 256 genes up in HH, 237 genes up in HR; RR vs HR, 177 genes up in HR, 84 genes up in RR, p. value < 0.05 for all comparisons). D Hypergeometric gene set enrichment analysis for the genes that were increased in either RR vs HH or HR vs HH. E Gene module framework divided by disease status and FcγRIIA genotype. Comparison columns show the difference in average module expression for SLE vs control separated by the respective genotype, with significant differences annotated by a gold star. F–H show gene module scores for the Tan, Yellow, and the Brown gene module respectively across our cohort when divided by disease status and FcγRIIA allele status