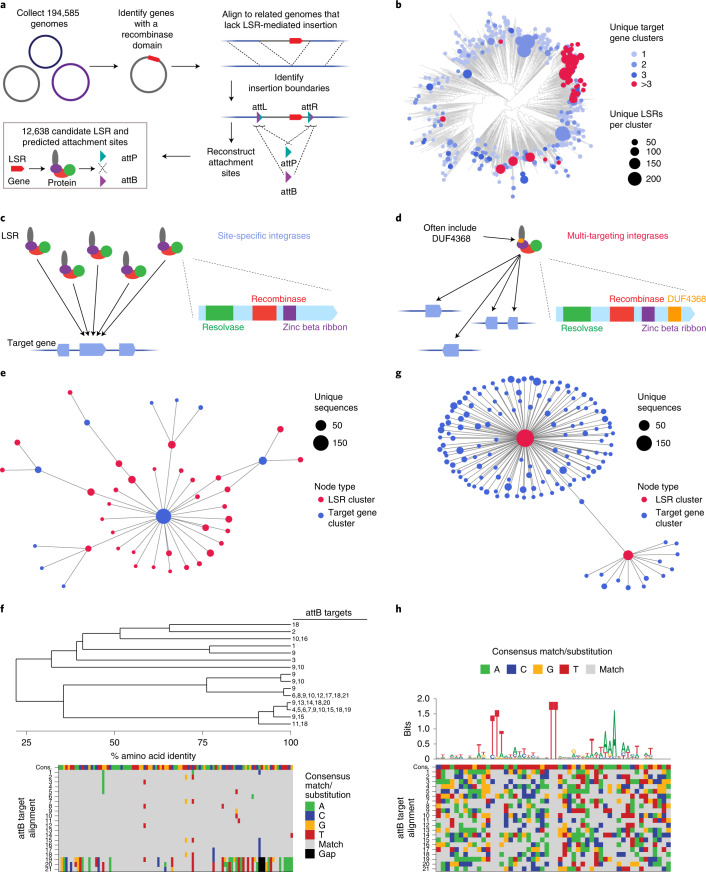
Fig. 1. Systematic discovery and classification of LSRs and their target site specificities.
a, Schematic of the computational workflow for systematic identification of LSRs and inference of their attachment sites. The gene harboring the recombinase domain is shown as a red rectangle. b, Phylogenetic tree of representative LSR orthologs clustered at 50% identity, annotated according to predicted target specificity of each LSR cluster. ‘Unique target gene clusters’ indicates the number of predicted target gene clusters, dots scaled to indicate the number of unique LSR sequences found in each LSR cluster. c, Schematic of the technique to identify site-specific LSRs that target a single gene cluster. The typical domain architecture of a site-specific LSR is illustrated. d, Schematic of the technique to identify multi-targeting LSRs. In brief, if a single cluster of related LSRs (clustered at 90% identity) integrates into multiple diverse target gene clusters (clustered at 50% identity), then the LSR cluster is considered multi-targeting. The typical domain architecture of a multi-targeting LSR is illustrated, commonly including a particular domain of unknown function, DUF4368. e, Example of an observed network of predicted site-specific LSRs found in our database. Each node indicates either an LSR cluster (red) or a target gene cluster (blue). Edges between nodes indicate that at least one member of the LSR cluster was found integrated into at least one member of the target gene cluster. f, Example of a hierarchical tree of diverse LSR sequences that target a set of closely related attB sequences. Numbers at the tip of the tree indicate the attB sequences in the alignment that are targeted by each LSR. Bottom is the alignment of related attB sequences. g, Example of an observed network of predicted multi-targeting LSRs. h, Schematic of an alignment of diverse attB sequences that are targeted by a single multi-targeting LSR. Each target sequence is aligned with respect to the core TT dinucleotide. Sequence logo above the alignment indicates conservation across target sites, a proxy for the sequence specificity of this particular LSR. The alignment is colored according to the consensus.