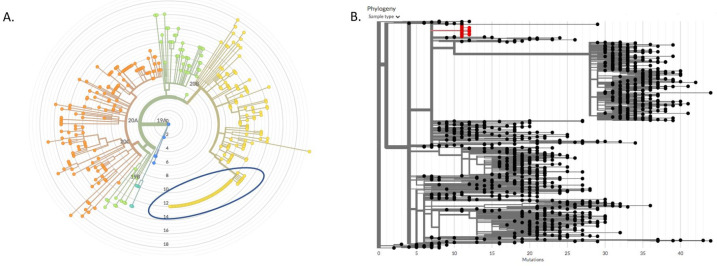
Figure 1.
Phylogeny of outbreak-associated strain of SARS-CoV-2 among participants in the Safe Campus Initiative. A. A maximum likelihood phylogeny constructed from 357 genomes sequenced by the Innovative Genomics Institute between May and July 2020 constructed using Nextstrain. Branch lengths represent divergence from Wuhan reference genome at center. Blue circle marks cluster of identical genomes from a campus super-spreader event. B. A 1,057 node subtree of a neighbor-joining tree constructed with all SARS-CoV-2 sequences to date (constructed using UShER with over 1 million genomes in April 2021), showing the most similar genomes to the super-spreader event cluster (in red). There are no descendant branches from the cluster, demonstrating that the outbreak was contained and the lineage died out.