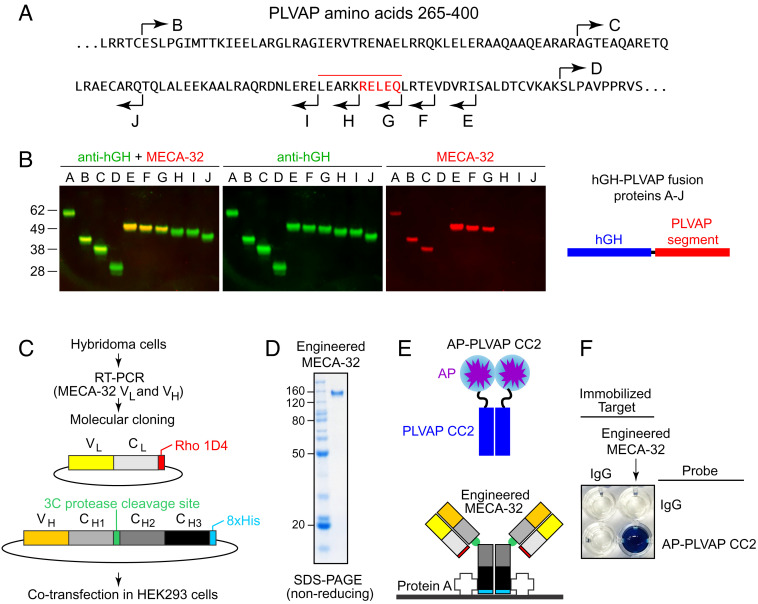
Fig. 9.
Mapping the epitope for MECA-32 on the PLVAP ECD. (A) The sequence of the PLVAP ECD between amino acids 265 and 400 showing the end points of deletions that remove progressively larger regions from the N terminus (rightward arrows B–D) and the end points of deletions that remove progressively larger regions from the C terminus (leftward arrows E–J). Amino acids essential for MECA-32 binding, as defined by PLVAP deletion analysis, are highlighted in red, with the overlying red bar indicating a larger region that likely encompasses the MECA-32 epitope. (B) SDS-PAGE and immunoblotting of hGH fusions with the PLVAP ECD deletions indicated in (A). Blots were probed with rabbit anti-hGH and mouse mAb MECA-32, and the primary antibodies were visualized with fluorescent secondary antibodies. Fusion proteins A-D extend to the C terminus of PLVAP. The N terminus of fusion protein A is at PLVAP amino acid 104 (TRREME...). Fusion proteins E-J have their N termini at PLVAP amino acid 143 (QEQLKE...). MECA-32 binds to fusion proteins A-C and E-G but not to fusion proteins D and H-J. (C) Workflow for MECA-32 Ab molecular cloning, construct design, and protein expression. (D) SDS-PAGE under nonreducing conditions of the engineered MECA-32 Ab (MECA-32/hIgG). (E) Diagram of the protein–protein interaction assay. MECA-32/hIgG was immobilized on protein A–coated wells. A fusion protein comprising alkaline phosphatase (AP) fused to PLVAP CC2 (AP–PLVAP CC2) was used as a probe. (F) A colorimetric AP reaction was used to detect the interaction between MECA-32/hIgG and AP–PLVAP CC2. Bovine IgG served as the negative control.