Abstract
Objective
To design a deep learning model based on multimodal magnetic resonance image (MRI) sequences for automatic parotid neoplasm classification, and to improve the diagnostic decision‐making in clinical settings.
Methods
First, multimodal MRI sequences were collected from 266 patients with parotid neoplasms, and an artificial intelligence (AI)‐based deep learning model was designed from scratch, combining the image classification network of Resnet and the Transformer network of Natural language processing. Second, the effectiveness of the deep learning model was improved through the multi‐modality fusion of MRI sequences, and the fusion strategy of various MRI sequences was optimized. In addition, we compared the effectiveness of the model in the parotid neoplasm classification with experienced radiologists.
Results
The deep learning model delivered reliable outcomes in differentiating benign and malignant parotid neoplasms. The model, which was trained by the fusion of T2‐weighted, postcontrast T1‐weighted, and diffusion‐weighted imaging (b = 1000 s/mm2), produced the best result, with an accuracy score of 0.85, an area under the receiver operator characteristic (ROC) curve of 0.96, a sensitivity score of 0.90, and a specificity score of 0.84. In addition, the multi‐modal paradigm exhibited reliable outcomes in diagnosing the pleomorphic adenoma and the Warthin tumor, but not in the identification of the basal cell adenoma.
Conclusion
An accurate and efficient AI based classification model was produced to classify parotid neoplasms, resulting from the fusion of multimodal MRI sequences. The effectiveness certainly outperformed the model with single MRI images or single MRI sequences as input, and potentially, experienced radiologists.
Level of Evidence
3 Laryngoscope, 133:327–335, 2023
Keywords: artificial intelligence, deep learning, machine learning, magnetic resonance imaging, neoplasms, parotid neoplasms
An accurate and efficient artificial intelligence‐based classification model was produced to classify parotid neoplasms, resulting from the fusion of multimodal magnetic resonance image (MRI) sequences. The effectiveness certainly outperformed the model with single MRI images or single MRI sequences as input, and potentially, experienced radiologists.
INTRODUCTION
Deep learning is a subdivision of artificial intelligence (AI). In recent years, the demand for deep learning in medical diagnosis has gradually emerged, largely due to the technological advancement in the pertaining field. A convolutional neural network (CNN) is the fundamental algorithm of deep learning. At present, CNN has led to respectful outcomes in the application of the two‐dimensional magnetic resonance image (MRI) image classification 1 , 2 , 3 , 4 and it was recently suggested that the CNN trained by dynamic contrast enhancement MRI sequence would result in improved performance on the classification and localization of breast cancer. 5
The tumors of the salivary gland mostly occur in the parotid gland, which is the largest salivary gland in the human body, and about 25% of parotid neoplasms are malignant. 6 , 7 , 8 Surgery is the primary treatment option for parotid neoplasms, and detailed evaluation is typically carried out before the surgery in order to select the appropriate surgical tactic for a particular pathological tumor type. The preoperative assessment of parotid gland tumors mainly includes medical history, physical examination, imaging examination, fine needle aspiration cytology (FNAC), and so on. 6 An FNAC is in general invasive and less diagnostically sensitive to malignant tumors. 9 , 10 However, other studies proposed that the hollow needle biopsy technique has high sensitivity and specificity for the diagnosis of parotid neoplasms, while the accuracy also heavily depends on the doctor's surgical skills and the selection of equipment. 11 The accuracy of preoperative FNAC can be effectively improved with the assistance of ultrasound, but as a stand‐alone preoperative evaluation method, the capability of ultrasound is limited. 12 Typically, the imaging methods for evaluating the parotid gland tumor include ultrasound, computerized tomography, and MRI. Comparing the first two methods, MRI is more effective and accurate to obtain the tumor's internal structural feature and spatial relationship with surrounding tissues. 13 Yet, further advancement in the functional MRI has recently been evident in terms of the diagnostic accuracy of disease. 7 , 14 , 15 For example, using a multiparametric non‐contrast MRI approach, Takumi et al. recently improved the differentiation of malignant salivary gland lesions from the benign ones. 16
In toto, MRI plays a pivotal role in the non‐invasive evaluation of parotid gland tumors, therefore, any strategy or paradigm that potentially improves the accuracy and efficiency of MRI‐based preoperative diagnosis will greatly benefit patients with parotid neoplasms. An AI‐based deep learning has been used to improve the performance of MRI in the diagnosis of parotid gland tumors. Matsuo et al. 17 combined the deep learning technique and an anomaly detection strategy, and applied the technology to a small and unbalanced MRI dataset of parotid tumors, resulting in a diagnostic performance superior to radiologists. The model achieved an area under the receiver operator characteristic curve (ROC‐AUC) of 0.86. Another study by Chang et al. 18 used multimodal MRI images to train deep learning models, as well as the transfer learning method, to classify parotid gland tumors. The model accurately distinguished the benign tumor subtypes of pleomorphic adenoma and Warthin tumor, but unfortunately with poor sensitivity in malignant tumor detection. The present study aimed to generate a deep learning model based on the multimodal MRI sequence to classify the parotid neoplasms automatically, so as to provide better support in clinical decision‐making.
MATERIALS AND METHODS
Patient Cohort and MRI Protocol
Participants
Inclusion criteria: Patients who underwent surgical resection of the parotid gland tumor from January 1st, 2015 to May 31st, 2020 in the Eye, Ear, Nose and Throat (EENT) Hospital of Fudan University, and the tumor was post‐surgically confirmed as parotid neoplasms through pathological assessment. In addition, the corresponding MRI examination was performed prior to the surgery.
Exclusion criteria: Patients with pathological assessment indicating schwannoma, eosinophilic adenoma, cystic degeneration, hemangioma or uncertain pathologies. Also excluded if the MRI examination was conducted without being contrast‐enhanced T1‐weighted (CE‐T1) or diffusion‐weighted (DWI).
Ethics: The ethical committee of the EENT Hospital approved this study (#2021176). As a retrospective study without any adverse effect on de‐identified subjects, the patient consent form was exempt.
Patient profile: The study included 266 patients (153 males and 113 females) with parotid neoplasms. The average age was 50.9, ranging from 15 to 85. There were 218 cases of benign tumors and 48 cases of malignant tumors. The benign tumors were further categorized into the subtypes of pleomorphic adenoma, Warthin tumor, and basal cell adenoma.
MRI protocol
All patients voluntarily underwent contrast‐enhanced MRI and DWI for the assessment of disease before surgery. We collected multiple MRI sequences, including T1‐weighted (T1), T2‐weighted (T2), postcontrast T1‐weighted (CE‐T1), apparent diffusion coefficient map (ADC), and DWI (b = 1000 s/mm2 and b = 0 s/mm2). Images were acquired by one of the two MRI 3.0T units (Magnetom Verio and Magnetom Prisma, Siemens Healthcare, Erlangen, Germany), with combined head and neck coils. Acquired images were stored and processed in the format of Digital Imaging and Communications in Medicine (DICOM).
The detailed MRI parameters were as follows. The field of view (FOV) was 240 × 240 mm, the matrix was 320 × 240, the thickness of axial images was 3–4 mm, and the thickness of coronal images was 3–4 mm. Axial fat‐suppressed T2‐weighted images were obtained from the turbo‐spin‐echo (TSE) sequence with TR/TE = 4460/78 ms. T1‐weighted images were also obtained from the TSE sequence with TR/TE = 849/11 ms. The gadolinium‐contrast‐enhanced fat‐suppressed T1 weighted images were acquired immediately with VIBE sequence, TR/TE = 3.73/1.52 ms, after a bolus of an intravenous injection of 0.1 mmol/kg using Gadoteridol injection (BIPSO GmbH, Germany) at a 2–3 ml/s rate followed by a 10 ml saline flush at the same rate with a power injector. The imaging parameters of RESOLVE DWI were as follows: TR/TE = 3600/65 ms, slice thickness/gap = 4/0.1 mm, FOV = 220 × 230 mm, matrix = 160 × 60 mm, voxel size = 2 × 2 × 4 mm, diffusion mode = 4 scan trace, b = 0 or 1000 s/mm2.
Data
Data calibration
Software “ITK‐SNAP” (www.itksnap.org) was used to calibrate MRI images. 19 All the slices containing tumors in the MRI sequences were calibrated by in‐house physicians and formed as the input sequence of the deep learning model, and the tumors were classified according to the pathology into four categories, namely, pleomorphic adenoma, Warthin tumor, basal cell adenoma, and malignant tumor. Later, the tumor was inclusively framed, as demonstrated within the CE‐T1 sequence (Fig. 1).
Fig. 1.
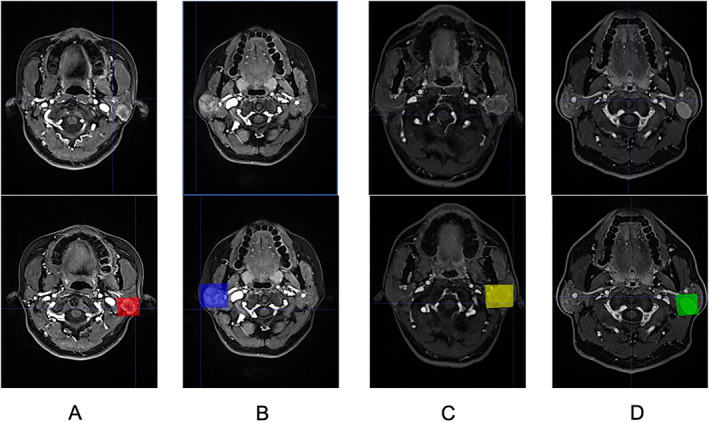
Illustrations of the calibrated magnetic resonance images in the dataset, each shaded rectangle represents the location of an identified parotid tumor. (A) malignant tumor, (B) pleomorphic adenoma, (C) basal cell adenoma, (D) Warthin tumor.
Dataset distribution
Randomly stratified sampling was carried out to keep equivalent proportions, that is, balanced, benign and malignant tumors between the training and the test sets. First, all data were divided into two layers based on benign versus malignant tumors. Then, the two layers of data were further split into a training set and a test set in the ratio of 4:1. In addition, 20% of the training set was used for validation (see Table I for detailed data distribution).
TABLE I.
Distribution of Pathological Subtypes of Parotid Tumors Between the Training Set and the Test Set.
| Data | Classification | Total | |
|---|---|---|---|
| Benign Tumor | Malignant Tumor | ||
| Training | 174 (102, 72) | 38 (19, 19) | 212 (121, 91) |
| Test | 44 (28, 16) | 10 (4, 6) | 54 (32, 22) |
| Total | 218 (130, 88) | 48 (23, 25) | 266 (153, 113) |
Numbers in the parentheses (male, female) indicate gender distribution.
Data preparation
Data augmentation
To achieve proper convergence, the deep learning model usually requires a huge amount of data during the model training process. However, images are unique in medical settings, because it is often unrealistic to obtain a large number of compatible and calibrated MRI images. Therefore, the original dataset needs to be expanded through data augmentation to improve the training process adequately. Here, using a series of data augmentation strategies, 20 we achieved a 10‐time randomized enhancement on the annotated images with calibration frames in each MRI sequence.
Data preprocessing
Typically, the CNN‐like Resnet takes RGB images as input, 21 that has three channels that capture red, green, and blue signals respectively. However, the MRI image generated in each modal is in grayscale with a single channel and cannot be directly processed by CNN. Therefore, we experimented with two different preprocessing methods to convert the MRI images to eligible CNN input format, to take full advantage of the pre‐trained networks. To briefly sum, the MRI images were resized to 224 × 224 according to the input requirement of Resnet‐18, then converted to RGB format in one of the two following methods.
In the single‐modal paradigm, the same MRI sequence served as reiterated input of RGB channels.
In the multi‐modal paradigm, compatible modalities were combined, such as T1, T2 and CE‐T1, so that each MRI sequence served as the channel input of individual RGB channels. 17
Deep Learning Paradigms
Single‐modal paradigm
Image classification network
Upon the model development, we firstly designed an image classification network of parotid gland tumors based on the Resnet‐18, as illustrated in Figure 2. The backbone of Resnet‐18 was initialized by pre‐trained weights on ImageNet, 22 given our dataset was not adequate to train a deep learning model effectively. The model was composed of the first two layers of Resnet18 and ROI‐Align 23 and extracted the tumor features using the MRI frame and the position of the bounding box as input. The classification output logits were calculated by the tumor features through the fully connected network, and the prediction was produced by the SoftMax function. During the training process, the cross‐entropy function was used to calculate the loss of the output and the actual label, and the loss was optimized by the ADAM optimizer with an initial learning rate 1e−4, originally proposed by Kingma and Ba. 24 In order to prevent overfitting, we also used p = 0.5 to perform a dropout operation on the fully connected layer. As mentioned above, the image classification network generated vectors representing image features. Two‐dimensional images from the six individual modals of MRI sequences were used as the training data.
Fig. 2.
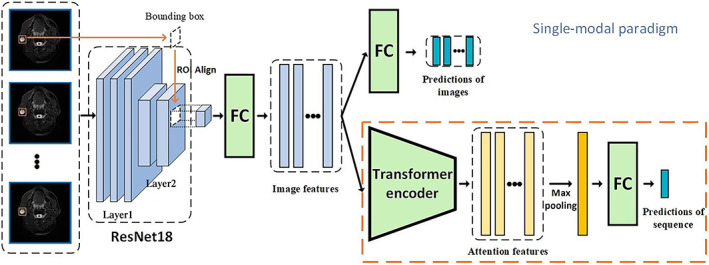
An illustration of the two‐stage single‐modal paradigm. In the first stage, an image classification network was trained to produce a vector describing image features through the first two layers of Resnet‐18 and the ROI‐Align. In the second stage, using the newly extracted image features as the input, a sequence classification network further extracted the sequential features and identified the subtype of tumors. FC = fully‐connected network.
Sequence classification network
In recent years, the Transformer network with a multi‐head self‐attention mechanism that learns multi‐space attention features from the sequence of original features has made remarkable achievements in the field of natural language processing and showed great promise in other sequence processing fields. 25 In the present study, we constructed the sequence classification model based on the Transformer network. The model utilized the pre‐trained image classification network to obtain tumor features that were extracted from an original MRI sequence and use the encoder structure in the Transformer network to extract self‐attention information. The number of encoder layers and number of heads were set to 1 and 4 respectively by cross‐validation. The encoder layer produced a self‐attention feature that contains sequential information. The features were max‐pooled along sequential dimensions to achieve the tumor feature of the entire input sequence. Finally, logits were calculated in the same way as in the image classification network. During the training process, the parameters of the image classification network were determined. The sequence classification network was optimized by the ADAM with an initial learning rate of 1e−5. We also used p = 0.5 to perform a dropout operation on the fully connected layer to avoid overfitting.
Multi‐modal paradigm
The multi‐modal paradigm, based on the sequence classification model, aimed to explore the improvement by the fusion of multiple modalities sequences. Specifically, the fusion operation was implemented inside a deep learning framework, namely, Pytorch (www.pytorch.org). We experimented with two methods of fusion, early fusion, and late fusion. In early fusion, as shown in Figure 3, the data of multiple modalities were only fused according to the RGB channel in the data preprocessing stage, 17 , 21 as the input of CNN. In late fusion, we used multiple CNN branches to extract features from single‐modal sequences. The feature vectors of different sequences, obtained in the above process, were fused as the input of the Transformer encoder with one of the following two methods. One method is the vector addition (“Add.”), where feature vectors were combined from multiple modalities to obtain a vector of the same dimension. The other method was vector splicing (“Spl.”), where feature vectors were concatenated to a higher dimension, maintaining the characteristics of each vector.
Fig. 3.
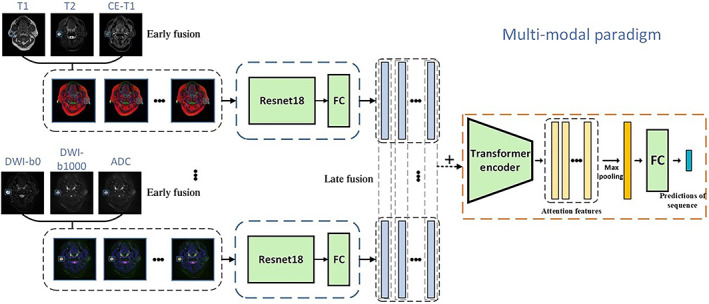
An example of the multi‐modal paradigm that combined early and late fusion methods. The model has two convolutional neural network branches that merged corresponding modalities together. Extracted features were subsequently combined as the input of the Transformer encoder. FC = fully‐connected network.
Cross‐Validation and Model Evaluation
During the training process, a five‐fold cross‐validation method was used to optimize hyperparameters. The patients in the training set were divided into five groups by stratified random sampling. To evaluate the proposed model comprehensively, we used accuracy, the area under the ROC curve (ROC‐AUC), sensitivity, and specificity as evaluation metrics. In addition, an experienced radiologist analyzed and scored the MRI sequences of all cases in the test dataset. He had 20+ years of experience in diagnostic radiology specializing in head and neck surgery, 10+ years of experience in MRI diagnosis, and 1000+ identifications of parotid neoplasms. Here, his diagnostic decision was based on the identical “deep learning” dataset without additional clinical information from the patients. He determined the tumor malignancy, severing as a benchmark to evaluate the model performance. Briefly, tumors were scored by a five‐point scale system, with 1 point being a definitive benign tumor, five indicating definitely malignant, and three being undetermined. A ROC curve was subsequently established based on the scoring system, evaluating the diagnostic power of the radiologist. 17 , 26
RESULTS
Single‐Modal Paradigm
Image classification network
The effectiveness of the deep learning model, for differentiating benign and malignant parotid gland tumors, trained by different input options of individual MRI images is shown in Table II. The AUC‐ROC score of the model, trained by CE‐T1 images, was the highest (0.85) by single‐image inputs. The best accuracy score of the model was 0.84, resulting from the training input of DWI‐b1000 images. The sensitivity of the training model was low in general, with the highest sensitivity of 0.68, obtained from DWI‐b0 images training input.
TABLE II.
The Performance Scores of Single‐Modal Paradigms and the Comparison Between Single‐Image and Single‐Sequence Inputs.
| Training Input | Accuracy | ROC‐AUC | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| T1 | Single‐image | 0.70 (0.65–0.75) | 0.75 (0.68–0.82) | 0.60 (0.48–0.71) | 0.73 (0.67–0.78) |
| Single‐sequence | 0.71 (0.57–0.84) | 0.75 (0.56–0.91) | 0.60 (0.22–0.89) | 0.73 (0.60–0.85) | |
| T2 | Single‐image | 0.78 (0.73–0.82) | 0.80 (0.73–0.86) | 0.60 (0.48–0.74) | 0.81 (0.77–0.86) |
| Single‐sequence | 0.84 (0.73–0.94) | 0.88 (0.75–0.98) | 0.71 (0.44–1) | 0.86 (0.75–0.98) | |
| CE‐T1 | Single‐image | 0.81 (0.78–0.85) | 0.85 (0.80–0.90) | 0.47 (0.35–0.59) | 0.89 (0.85–0.93) |
| Single‐sequence | 0.87 (0.78–0.96) | 0.89 (0.77–0.98) | 0.70 (0.44–1) | 0.91 (0.80–1) | |
| DWI‐b0 | Single‐image | 0.75 (0.63–0.88) | 0.82 (0.64–0.95) | 0.68 (0.33–0.89) | 0.76 (0.63–0.90) |
| Single‐sequence | 0.76 (0.63–0.88) | 0.83 (0.68–0.95) | 0.79 (0.56–1) | 0.75 (0.63–0.88) | |
| DWI‐b1000 | Single‐image | 0.84 (0.80–0.88) | 0.82 (0.75–0.88) | 0.58 (0.46–0.72) | 0.90 (0.86–0.93) |
| Single‐sequence | 0.84 (0.73–0.94) | 0.89 (0.78–0.98) | 0.80 (0.56–1) | 0.84 (0.73–0.95) | |
| ADC | Single‐image | 0.82 (0.78–0.86) | 0.77 (0.69–0.84) | 0.43 (0.29–0.58) | 0.90 (0.87–0.94) |
| Single‐sequence | 0.78 (0.65–0.88) | 0.82 (0.68–0.94) | 0.60 (0.33–0.89) | 0.82 (0.70–0.93) | |
Values in the parentheses indicate 95% confidence interval. Notable values are in bold, see main text for details.
ADC = apparent diffusion coefficient; DWI = diffusion‐weighted; ROC = receiver operator characteristic.
Sequence classification network
According to the result based on single‐image input, we concluded that the MRI images of CE‐T1, T2, DWI‐b0, and DWI‐b1000 were more effective to differentiate benign and malignant tumors. Thus, we subsequently conducted CNN training with various MRI sequences. The performance scores with a 95% confidence interval from six different inputs are presented in Table II. The overall performance of the model trained by MRI sequences, was largely better than the corresponding single‐image input, except for the T1 sequence. The AUC‐ROC of the model, trained by the CE‐T1 sequence, reached an equivalent performance to that of the DWI‐b1000 sequence, with a score of 0.89, superior to other sequence inputs. Additionally, the best sensitivity score was achieved by the DWI‐b1000 input, the best specificity by the CE‐T1 input, and the best accuracy by the CE‐T1 input.
Multi‐Modal Paradigm
In real‐world practice, clinicians and radiologists evaluate parotid gland tumors through multiple MRI sequences. The present study simulated the practice and used multiple sequences as the training input of the network to extract features from multi‐modal MRI sequences (Table III). The fusion of multiple modal MRI images evidently elevated the efficiency of the deep learning diagnosis model, with significantly improved accuracy, ROC‐AUC, sensitivity, and specificity scores. The combination of MRI sequences in the multi‐modal paradigm enhanced ROC curves from that of individual sequences in the single‐modal paradigm (Fig. 4A–D). In addition, the late‐fusion strategy resulted in better effectiveness than the early‐fusion strategy, while the vector splicing method exhibited a superior outcome to the vector addition method. In sum, the deep learning model trained by T2, CE‐T1, and DWI‐b1000, using the vector splicing method with a late‐fusion strategy, presented the best performance, with an AUC‐ROC score of 0.96, accuracy of 0.85, sensitivity of 0.90, and specificity of 0.84 (Table III). It is also worth noting that the best result was not achieved by the fusion of all the modalities of MRI sequences.
TABLE III.
The Performance Scores of the Multi‐Modal Deep Learning Model With Different Fusion Strategies and an Experienced Radiologist.
| Training Input | Vector | Accuracy | ROC‐AUC | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
|
T1 T2 CE‐T1 |
Early‐fusion | 0.83 (0.73–0.92) | 0.88 (0.74–0.98) | 0.89 (0.67–1) | 0.82 (0.70–0.93) | |
| Late‐fusion | Add. | 0.87 (0.78–0.96) | 0.88 (0.75–1) | 0.69 (0.33–1) | 0.91 (0.80–1) | |
| Spl. | 0.85 (0.76–0.94) | 0.89 (0.72–0.99) | 0.70 (0.44–1) | 0.89 (0.78–0.98) | ||
|
T2 CE‐T1 DWI‐b1000 |
Early‐fusion | 0.76 (0.63–0.88) | 0.86 (0.74–0.96) | 0.70 (0.44–1) | 0.77 (0.63–0.90) | |
| Late‐fusion | Add. | 0.85 (0.73–0.94) | 0.93 (0.83–0.99) | 0.80 (0.56–1) | 0.86 (0.75–0.95) | |
| Spl. | 0.85 (0.75–0.93) | 0.96 (0.89–1) | 0.90 (0.67–1) | 0.84 (0.73–0.95) | ||
|
DWI‐b0 DWI‐b1000 ADC |
Early‐fusion | 0.82 (0.69–92) | 0.83 (0.65–0.96) | 0.60 (0.22–0.89) | 0.86 (0.75–0.98) | |
| Late‐fusion | Add. | 0.83 (0.71–0.94) | 0.87 (0.67–1) | 0.79 (0.56–1) | 0.84 (0.73–0.95) | |
| Spl. | 0.94 (0.88–0.1) | 0.91 (0.7–1) | 0.89 (0.67–1) | 0.95 (0.88–1) | ||
| All MRI sequences | (Early + late)‐fusion | Add. | 0.78 (0.65–0.88) | 0.76 (0.56–0.91) | 0.59 (0.33–0.89) | 0.82 (0.70–0.93) |
| Spl. | 0.76 (0.63–0.88) | 0.84 (0.70–0.96) | 0.69 (0.44–1) | 0.77 (0.65–0.90) | ||
| Late‐fusion | Add. | 0.89 (0.80–0.96) | 0.80 (0.56–1) | 0.70 (0.33–1) | 0.93 (0.85–1) | |
| Spl. | 0.81 (0.69–0.92) | 0.95 (0.86–1) | 0.81 (0.56–1) | 0.81 (0.70–0.93) | ||
| Radiologist | 0.70 (0.58–0.82) | 0.74 (0.59–0.86) | 0.70 (0.44–1) | 0.70 (0.58–0.85) | ||
Values in the parentheses indicate 95% confidence interval. Notable values are in bold, see main text for details. “Add.” = vector addition method, “Spl.” = vector splicing method.
ADC = apparent diffusion coefficient; DWI = diffusion‐weighted; MRI = magnetic resonance image; ROC = receiver operator characteristic.
Fig. 4.
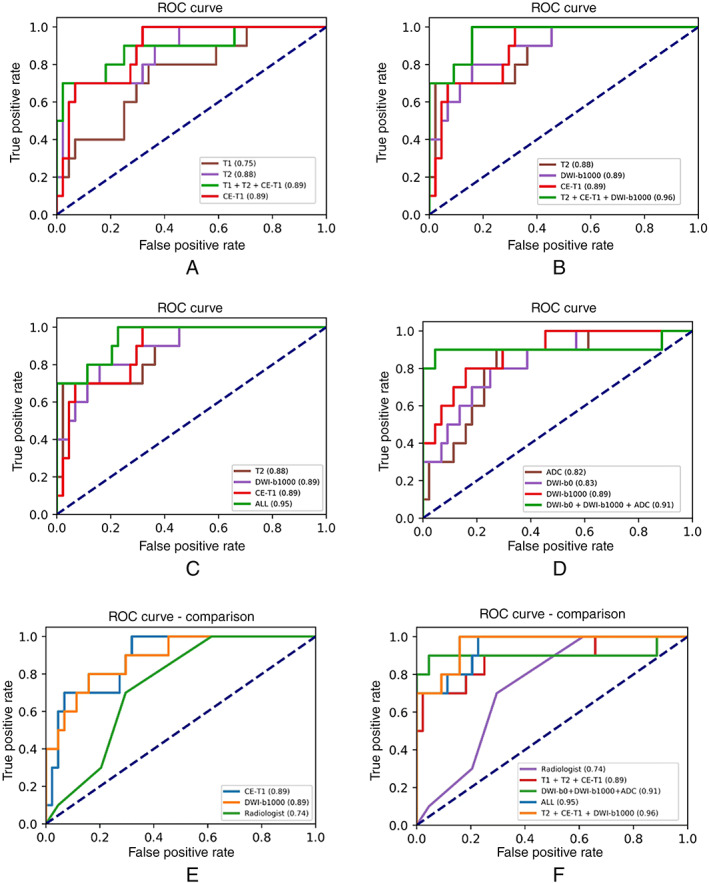
Receiver operator characteristic (ROC) curves of the multi‐modal deep learning paradigm and the corresponding single‐sequence ROC curves. (A) Multi‐model sequences (T1 + T2 + CE‐T1) and the corresponding single sequences, (B) Multi‐model sequences (T2 + CE‐T1 + DWI‐b1000), (C) Multi‐model sequences (all sequences), (D) Multi‐model sequences (DWI‐b0 + DWI‐b1000 + ADC). (E) Selected single‐modal ROC curves compared to a radiologist, (F) Multi‐modal ROC curves compared to the radiologist. ADC = apparent diffusion coefficient; DWI = diffusion‐weighted.
Performance Comparison Between the Deep Learning Model and Radiologists
The effectiveness of the present model trained by multi‐modal MRI sequences was compared to an experienced radiologist (Table III), ROC‐AUC curves from both optimally trained multi‐modal paradigm and single‐modal paradigm were also compared to the radiologist (Fig. 4E,F). Using either multi‐modal sequences or a single sequence as the training input, the model produced efficacious ROC curves, indicating superior diagnostic performance compared to the experienced radiologist.
The Effectiveness of the Multi‐Modal Paradigm to Distinguish Other Tumors
The above results indicated that the fusion of multi‐modal sequences improved the diagnostic performance of deep learning, specifically, in differentiating benign and malignant parotid tumors. The present study additionally applied the aforementioned, optimized methods to identify tumor subtypes, including pleomorphic adenoma, Warthin tumor, and basal cell adenoma. Here, using the fusion of all sequences produced the best performance in distinguishing the three tumor subtypes (Table IV). The best model performance was observed in the diagnosis of pleomorphic adenoma, with the best accuracy score of 0.93, ROC‐AUC of 0.96, the sensitivity of 0.96, and specificity of 0.90. The diagnostic effectiveness of the basal cell adenoma was considerably worse.
TABLE IV.
The Performance Scores of the Multi‐Modal Paradigm to Distinguish the Tumor Subtypes, Pleomorphic Adenoma (PMA), Warthin Tumor (WT), and Basal Cell Adenoma (BCA).
| Subtype | Training Input | Accuracy | ROC‐AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|
| PMA | T1/T2/CE‐T1 | 0.84 (0.73–0.92) | 0.95 (0.89–0.99) | 0.87 (0.71–1) | 0.81 (0.64–0.93) |
| ADC/DWI‐b0/b1000 | 0.78 (0.65–0.88) | 0.87 (0.77–0.96) | 0.83 (0.67–0.95) | 0.74 (0.57–0.89) | |
| All MRI sequences | 0.93 (0.86–0.98) | 0.96 (0.90–1) | 0.96 (0.86–1) | 0.90 (0.79–1) | |
| WT | T1/T2/CE‐T1 | 0.91 (0.82–0.98) | 0.93 (0.83–1) | 0.80 (0.62–1) | 0.95 (0.86–1) |
| ADC/DWI‐b0/b1000 | 0.87 (0.78–0.96) | 0.96 (0.91–1) | 0.87 (0.69–1) | 0.87 (0.75–0.97) | |
| All MRI sequences | 0.87 (0.78–0.96) | 0.97 (0.92–1) | 0.87 (0.69–1) | 0.92 (0.8–0.97) | |
| BCA | T1/T2/CE‐T1 | 0.78 (0.67–0.88) | 0.77 (0.59–0.95) | 0.33 (0–0.80) | 0.83 (0.73–0.93) |
| ADC/DWI‐b0/b1000 | 0.76 (0.63–0.88) | 0.77 (0.53–0.95) | 0.67 (0.2–1) | 0.77 (0.64–0.89) | |
| All MRI sequences | 0.73 (0.59–0.84) | 0.84 (0.69–0.96) | 0.84 (0.40–1) | 0.71 (0.57–0.84) |
Values in the parentheses indicate 95% confidence interval. Notable values are in bold, see main text for details.
ADC = apparent diffusion coefficient; DWI = diffusion‐weighted; MRI = magnetic resonance image; ROC = receiver operator characteristic.
DISCUSSION
The features of MRI sequences are critical in the diagnosis of parotid neoplasms. 27 , 28 , 29 , 30 The present study is innovative in terms of utilizing a newly designed CNN to classify parotid gland tumors. Briefly, the first two layers of the pre‐trained Resnet‐18 were selected to extract the overall image features based on the concept of learning transfer. Then, the tumor features were extracted by using ROI‐Align to match the position of the tumor's bounding box with the image features. Previous studies on the MRI‐based classification of parotid gland tumors are very limited. The present study for the first time applied the multiple MRI sequences to classify parotid neoplasms, and successfully outperformed the prior studies in which only individual MRI images were considered. Matsuo et al. 17 investigated several CNNs, including VGG16, MobileNet, Resnet‐50, and CVAE. The VGG16‐based model with the L2‐constrained SoftMax loss function resulted in the best diagnostic accuracy. Chang et al. 18 applied the U‐Net to the classification of parotid gland tumors, effectively detecting Warthin tumor and pleomorphic adenoma, but the model was not sensitive to the detection of malignant tumors. Xia et al. 31 also selected Resnet to distinguish benign and malignant tumors, but the tumor subtype detection was based on a simple decision tree structure.
The MRI sequences were processed by the pre‐trained CNN, resulting in a sequence of tumor features. Meanwhile, the Transformer network, which has achieved remarkable performance in the field of natural language processing, was used to obtain the features of the whole sequence through a multi‐head self‐attention mechanism. Moreover, this study integrated different modalities of MRI sequences and proposed a multi‐modal paradigm with various fusion strategies. Due to the limited dataset, overfitting in the deep learning model inevitably occurred. To resolve the issue, a two‐stage training method was adopted to optimize the model. That is, the image feature extraction network was initially trained, followed by the optimization of the sequence classification network. In addition, CNN and Transformer networks were simplified to avoid overfitting.
Certain selection biases existed in the retrospective study, for instance, some patients who did not undergo MRI examination were excluded. The present model can be further verified using external datasets from other hospitals. The model will especially benefit from a multicenter effort with expanded malignant cases, which were limited to 48 in the present study, and with considerable heterogeneity.
The superior performance of the present model was the result of collective effort based on continuous optimization and improvement by focusing on, for instance, the training input and feature extraction strategies. For future studies, the research team plan to enroll more patients with parotid tumors of different pathological subtypes and emphasis the classification of malignant tumors. Additionally, a semi‐supervised learning method can be used to generalize the model, by using additional unannotated images to minimize the likelihood of overfitting. Furthermore, a new approach 32 , 33 has been established to synthesize annotated MRI images from CT images for the purpose of image segmentation or data augmentation. The approach can be integrated into the present model with our in‐house CT dataset, resulting in an expanded training dataset, and subsequently, improved accuracy of the deep learning. With a greater training dataset, either by the recruitment of new patients or by artificial data augmentation strategies, we expect to use the deep learning model to facilitate clinical decision‐making on a broader patient cohort.
CONCLUSION
In the present study, an AI‐based deep learning model was established by selecting either individual MRI images or MRI sequences as the training input, using the reconstructed Resnet‐18 network to extract image features, and characterize the features through the Transformer network. Data fusion strategies were explored using various combinations of MRI sequences. In sum, a deep learning classification model based on the fusion of multi‐modal features was produced and successfully improved the artificial diagnostic efficiency resulting in superior performance over experienced imaging physicians. Along the course of the model development, the model performance has been continuously improved by optimizing the training input and data fusion methods, such as image sequence versus single‐image, multi‐modal versus single‐modal, late fusion versus early fusion, etc.
ACKNOWLEDGEMENT
This material is the result of work that was partly supported by the Office of Research & Development of Veterans Health Association No. RX002813. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Veterans Affairs or the United States Government.
Editor's Note: This Manuscript was accepted for publication on April 12, 2022.
Xu Liu, Yucheng Pan, Xin Zhang, and Yongfang Sha contributed equally to this study.
The authors have no financial relationships or conflicts of interest to disclose.
Contributor Information
Hongzhe Li, Email: hongzhe.li@va.gov, Email: hongzhe@gmail.com.
Jianping Liu, Email: jiulu123@sina.com.cn.
References
- 1. Schelb P, Kohl S, Radtke JP, et al. Classification of cancer at prostate MRI: deep learning versus clinical PI‐RADS assessment. Radiology. 2019;293:607‐617. [DOI] [PubMed] [Google Scholar]
- 2. Quon JL, Bala W, Chen LC, et al. Deep learning for pediatric posterior fossa tumor detection and classification: a multi‐institutional study. Am J Neuroradiol. 2020;41:1718‐1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hamm CA, Wang CJ, Savic LJ, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi‐phasic MRI. Eur Radiol. 2019;29:3338‐3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xi IL, Zhao Y, Wang R, et al. Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin Cancer Res. 2020;26:1944‐1952. [DOI] [PubMed] [Google Scholar]
- 5. Zhou J, Luo LY, Dou Q, et al. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging. 2019;50:1144‐1151. [DOI] [PubMed] [Google Scholar]
- 6. Moore MG, Yueh B, Lin DT, Bradford CR, Smith RV, Khariwala SS. Controversies in the workup and surgical management of parotid neoplasms. Otolaryngol Head Neck Surg. 2021;164:27‐36. [DOI] [PubMed] [Google Scholar]
- 7. Gökçe E. Multiparametric magnetic resonance imaging for the diagnosis and differential diagnosis of parotid gland tumors. J Magn Reson Imaging. 2020;52:11‐32. [DOI] [PubMed] [Google Scholar]
- 8. Abdel Razek AAK, Mukherji SK. State‐of‐the‐art imaging of salivary gland tumors. Neuroimaging Clin N Am. 2018;28:303‐317. [DOI] [PubMed] [Google Scholar]
- 9. Lameiras AR, Estibeiro H, Montalvão P, Magalhães M. Diagnostic accuracy and utility of fine‐needle aspiration cytology in therapeutic management of parotid gland tumours. Acta Otorrinolaringol Esp. 2019;70:74‐79. [DOI] [PubMed] [Google Scholar]
- 10. Shkedy Y, Alkan U, Mizrachi A, et al. Fine‐needle aspiration cytology for parotid lesions, can we avoid surgery? Clin Otolaryngol. 2018;43:632‐637. [DOI] [PubMed] [Google Scholar]
- 11. Cengiz AB, Tansuker HD, Gul R, Emre F, Demirbas T, Oktay MF. Comparison of preoperative diagnostic accuracy of fine needle aspiration and core needle biopsy in parotid gland neoplasms. Eur Arch Otorhinolaryngol. 2021;278:4067‐4074. [DOI] [PubMed] [Google Scholar]
- 12. Lanišnik B, Levart P, Čizmarevič B, Švagan M. Surgeon‐performed ultrasound with fine‐needle aspiration biopsy for the diagnosis of parotid gland tumors. Head Neck. 2021;43:1739‐1746. [DOI] [PubMed] [Google Scholar]
- 13. Liu Y, Zheng J, Lu X, et al. Radiomics‐based comparison of MRI and CT for differentiating pleomorphic adenomas and Warthin tumors of the parotid gland: a retrospective study. Oral Surg Oral Med Oral Pathol Oral Radiol. 2021;131:591‐599. [DOI] [PubMed] [Google Scholar]
- 14. Elmokadem AH, Abdel Khalek AM, Abdel Wahab RM, et al. Diagnostic accuracy of multiparametric magnetic resonance imaging for differentiation between parotid neoplasms. Can Assoc Radiol J. 2019;70:264‐272. [DOI] [PubMed] [Google Scholar]
- 15. Maraghelli D, Pietragalla M, Cordopatri C, et al. Magnetic resonance imaging of salivary gland tumours: key findings for imaging characterisation. Eur J Radiol. 2021;139:109716. [DOI] [PubMed] [Google Scholar]
- 16. Takumi K, Nagano H, Kikuno H, Kumagae Y, Fukukura Y, Yoshiura T. Differentiating malignant from benign salivary gland lesions: a multiparametric non‐contrast MR imaging approach. Sci Rep. 2021;11:2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsuo H, Nishio M, Kanda T, et al. Diagnostic accuracy of deep‐learning with anomaly detection for a small amount of imbalanced data: discriminating malignant parotid tumors in MRI. Sci Rep. 2020;10:19388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang YJ, Huang TY, Liu YJ, Chung HW, Juan CJ. Classification of parotid gland tumors by using multimodal MRI and deep learning. NMR Biomed. 2021;34:e4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yushkevich PA, Piven J, Hazlett HC, et al. User‐guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116‐1128. [DOI] [PubMed] [Google Scholar]
- 20. Bloice MD, Roth PM, Holzinger A. Biomedical image augmentation using Augmentor. Bioinformatics. 2019;35:4522‐4524. [DOI] [PubMed] [Google Scholar]
- 21. He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, 2016. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, 770‐778. [Google Scholar]
- 22. Deng J, Dong W, Socher R, Li LJ, Li FF. ImageNet: a large‐scale hierarchical image database. 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR 2009), 20–25 June 2009, Miami, Florida, USA; 2009.
- 23. Mask R‐CNN . IEEE International Conference on Computer Vision (ICCV); 2017.
- 24. Kingma DP, Ba JL. Adam: a method for stochastic optimization. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- 25. Vaswani A, Shazeer N, Parmar N, Uszkoreit Jakob, Jones Llion, Gomez Aidan N., Kaiser Łukasz, Polosukhin Illia Attention is all you need. 31st Annual Conference on Neural Information Processing Systems (NIPS); 2017 Dec 04–09; Long Beach, CA; 2017.
- 26. Nikpanah M, Xu Z, Jin D, et al. A deep‐learning based artificial intelligence (AI) approach for differentiation of clear cell renal cell carcinoma from oncocytoma on multi‐phasic MRI. Clin Imaging. 2021;77:291‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masmoudi M, Hasnaoui M, Guizani R, Lahmar R, Jerbi S, Mighri K. Performance of the magnetic resonance imaging in parotid gland tumor histopathology. Pan Afr Med J. 2021;39:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Liu S, Tang Y, et al. Performance of diffusion‐weighted imaging for the diagnosis of parotid gland malignancies: a meta‐analysis. Eur J Radiol. 2021;134:109444. [DOI] [PubMed] [Google Scholar]
- 29. Nardi C, Tomei M, Pietragalla M, et al. Texture analysis in the characterization of parotid salivary gland lesions: a study on MR diffusion weighted imaging. Eur J Radiol. 2021;136:109529. [DOI] [PubMed] [Google Scholar]
- 30. Wei PY, Shao C, Huan T, Wang HB, Ding ZX, Han ZJ. Diagnostic value of maximum signal intensity on T1‐weighted MRI images for differentiating parotid gland tumours along with pathological correlation. Clin Radiol. 2021;76:472.e19‐472.e25. [DOI] [PubMed] [Google Scholar]
- 31. Xia X, Feng B, Wang J, et al. Deep learning for differentiating benign from malignant parotid lesions on MR images. Front Oncol. 2021;11:632104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lei Y, Wang T, Tian S, et al. Male pelvic multi‐organ segmentation aided by CBCT‐based synthetic MRI. Phys Med Biol. 2020;65:035013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kieselmann JP, Fuller CD, Gurney‐Champion OJ, Oelfke U. Cross‐modality deep learning: contouring of MRI data from annotated CT data only. Med Phys. 2021;48:1673‐1684. [DOI] [PMC free article] [PubMed] [Google Scholar]


