TO THE EDITOR
The opportunistic pathogen Staphylococcus aureus accounts for the majority of all skin and soft tissue infections in the United States and infects 30—100% of atopic dermatitis (AD) lesions (Geoghegan et al., 2018; Parlet et al., 2019). Conversely, S. aureus only transiently colonizes healthy skin, suggesting that this pathogen may be actively excluded from this environment (Geoghegan et al., 2018). Healthy human skin is home to a diverse community of normal flora, including the coagulase-negative staphylococci (CoNS), and many CoNS directly compete with S. aureus through a variety of mechanisms to maintain skin homeostasis and colonization resistance (Parlet et al., 2019).
One mechanism of CoNS colonization resistance is quorum-sensing interference. All staphylococci encode the conserved accessory gene regulator (agr) quorum-sensing system, which senses and responds to its cognate autoinducing peptide (AIP) signal in a cell density–dependent manner (Thoendel et al., 2011). S. aureus agr is required for skin infection and degradation of the epithelium in AD because most virulence factors, including proteases, lipases, and toxins, are under agr control (Nakamura et al., 2020; Thoendel et al., 2011). S. aureus encodes four agr allelic variants, determined by a hypervariable region spanning agrBDC. Each variant senses and responds to a unique cognate AIP but can be inhibited by noncognate AIPs through intraspecies or interspecies cross-talk (Thoendel et al., 2011). The ubiquitous CoNS skin commensals S. hominis and S. epidermidis make AIPs that inhibit S. aureus agr and mitigate infection (Otto et al., 2001; Williams et al., 2019). However, many other healthy skin CoNS remain understudied. We hypothesize that these common skin colonizers may also play a role in maintaining colonization resistance through quorum sensing interference.
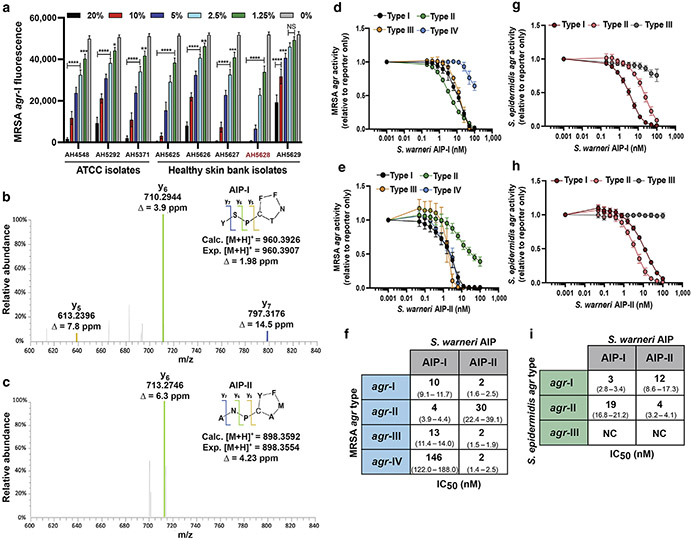
Staphylococcus warneri can be isolated from the head, nares, arms, legs, and feet of healthy individuals, but little is understood about its contributions to colonization resistance (Byrd et al., 2018; Kloos and Schleifer, 1975). We found that conditioned media (CM) from healthy skin isolates of S. warneri–inhibited S. aureus agr-I signaling in a dose-dependent manner, with no effect on growth (Figure 1a and Supplementary Figure S1). Sequence analysis of the AIP-encoding region (agrD) of each S. warneri strain revealed two putative agr types. The S. warneri AIP-I structure was previously identified (Gless et al., 2019), but to our knowledge, S. warneri agr allelic variation has not been previously reported. Liquid chromatography-mass spectrometry analysis of CM from an agr-I strain (AH4548) confirmed the previously published structure of AIP-I to be YSPc[CTNFF] (Gless et al., 2019), with m/z of 960.3907 (Figure 1b and Supplementary Figure S2). Liquid chromatography-mass spectrometry of CM from the agr-II strain (AH5628) revealed an eight amino acid peptide (ANPc[CAMFY]) with a measured m/z value for the [M+H]+ ion of 898.3592 (Figure 1c and Supplementary Figure S3).
Figure 1. S. warneri AIPs inhibit agr quorum sensing in opportunistic staphylococcal pathogens.
(a) The MRSA agr-I P3::YFP reporter strain was incubated with increasing doses of S. warneri cell-free CM from healthy skin or ATCC (Manassas, VA) isolates. The representative S. warneri agr-II strain is highlighted in red. Significant differences to 0% CM were calculated by ordinary one-way ANOVA followed by Dunnett’s MCT. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.0001. (b) MS confirmation of S. warneri AIP-I from CM. (c) MS identification of S. warneri AIP-II from CM. (d) MRSA agr-I-IV P3::YFP reporter strains incubated with increasing concentrations of synthetic S. warneri AIP-I. (e) MRSA agr-I-IV P3::YFP reporter strains incubated with increasing concentrations of synthetic S. warneri AIP-II. (f) Calculated IC50 values for synthetic AIP inhibition of MRSA quorum sensing. The 95% confidence intervals are reported in parenthesis. (g) S. epidermidis agr-I-III P3::sGFP reporters incubated with increasing concentrations of synthetic S. warneri AIP-I. (h) S. epidermidis agr-I-III P3::sGFP reporter strains incubated with increasing concentrations of synthetic S. warneri AIP-II. (i) Calculated IC50 values for synthetic AIP inhibition of S. epidermidis quorum sensing. The 95% confidence intervals are reported in parenthesis. The 24-hour fluorescence point is shown for a, d, e, g, and h. For all experiments, error bars are mean ± SD, and results are pooled from three independent experiments. IC50 was calculated with a four-parameter logistic regression curve. agr, accessory gene regulator; AIP, autoinducing peptide; Calc., calculated; CM, conditioned media; Exp., expected; IC50, half-maximal inhibitory concentration; MCT, multiple comparisons test; MRSA, methicillin-resistant Staphylococcus aureus; MS, mass-spectrometry; NS, not significant.
S. aureus agr function is critical for skin infection and exacerbation of AD lesions (Geoghegan et al., 2018; Nakamura et al., 2020). To determine whether S. warneri AIP-I (Figure 1d) or AIP-II (Figure 1e) could inhibit all of the four S. aureus agr classes, we treated fluorescent methicillin–resistant S. aureus (MRSA) agr reporters (P3::YFP) with increasing doses of each synthetic peptide. Both AIPs had half-maximal inhibitory concentration values in the low nanomolar range against most MRSA agr types, except for AIP-I, which weakly inhibited MRSA agr-IV (Figure 1f). Notably, AIP-II had potent activity (half-maximal inhibitory concentration, 4 nM) against MRSA agr-IV (Figure 1f).
S. epidermidis can also expand in AD lesions, and the S. epidermidis agr–regulated protease EcpA was shown to degrade the AD barrier (Byrd et al., 2017; Cau et al., 2021). However, only one study has identified inter-CoNS AIP-mediated cross-talk (Cau et al., 2021). Using fluorescent S. epidermidis reporters (P3::sGFP), we found that synthetic AIP-I (Figure 1g) and AIP-II (Figure 1h) were potent inhibitors of S. epidermidis agr-I, which is the most common S. epidermidis agr type on healthy and AD skin (Olson et al., 2014; Williams et al., 2019). Both AIPs inhibited agr-II with low nanomolar half-maximal inhibitory concentration but neither inhibited agr-III (Figure 1i). Together, our in vitro data suggest that competition between S. warneri and S. epidermidis or S. aureus may dampen the production of agr-regulated virulence factors.
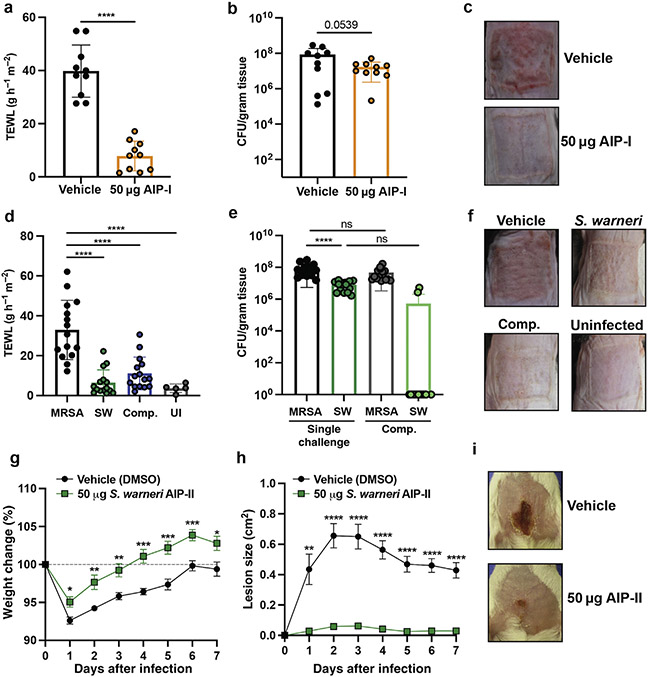
Given the potent activity of both S. warneri AIPs in vitro, we determined whether they might also be protective against MRSA-mediated skin damage in vivo. A total of 50 μg of synthetic S. warneri AIP-I dampened MRSA agr signaling and maintained barrier integrity as assessed by transepithelial water loss (Figure 2a), with no change in recovered bacteria (Figure 2b) in a mouse model of epicutaneous infection. This finding is similar to our in vitro findings that AIPs inhibited MRSA agr activation with no effect on growth. AIP-I application also inhibited erythema, redness, and scaling 72 hours after infection (Figure 2c). We then focused on the characterization of the novel S. warneri AIP-II, which had even greater inhibitory activity in vitro than AIP-I. In the epicutaneous model, coinoculation with equivalent numbers of an S. warneri AIP-II–producing strain and MRSA significantly decreased skin transepithelial water loss compared to MRSA alone (Figure 2d). No significant differences in MRSA numbers were recovered between the single and coinfection groups (Figure 2e). Coinfection with S. warneri also prevented barrier erythema and scaling (Figure 2f). Thus, S. warneri and its AIPs may contribute to the repression of S. aureus virulence factor production on the skin.
Figure 2. SW protects murine skin from MRSA damage.
(a) TEWL at 72 hpi with vehicle (DMSO) or 50 μg SW synthetic AIP-I (n = 10). Error bars are mean ± SD. (b) MRSA CFUs from vehicle or AIP-I treatment 72 hpi. Significant differences from MRSA alone (vehicle) were analyzed by ordinary one-way ANOVA followed by Dunnet’s MCT for TEWL or Tukey’s MCT for CFUs. ****P < 0.0001. (c) Representative images of back skin 72 hpi. (d) TWEL at 72 hpi for indicated groups (n = 15). (e) CFUs recovered from indicated groups 72 hpi (n = 15). Significant differences from MRSA alone (vehicle) were analyzed by ordinary one-way ANOVA followed by Dunnet’s MCT for TEWL or Tukey’s MCT for CFU data. ****P < 0.0001. (f) Representative images of back skin 72 hpi with indicated groups. (g) A 7-day weight change for groups administered vehicle (DMSO) or 50 μg synthetic SW AIP-II (n = 11 for vehicle, n = 10 for AIP). (h) A 7-day lesion-size change for groups administered vehicle (DMSO) or 50 μg synthetic SW AIP-II. Significant differences between groups were analyzed by multiple unpaired Student’s t-tests with FDR correction. *P < 0.05, **P < 0.005, and ***P < 0.0005. (i) Representative images at 3 dpi for indicated groups. AIP, autoinducing peptide; CFU, colony-forming unit; Comp., competition; FDR, false discovery rate; hpi, hour post infection; MCT, multiple comparisons test; MRSA, methicillin-resistant Staphylococcus aureus; ns, not significant; TEWL, transepithelial water loss; SW, Staphyloccocus warneri; UI, uninfected.
To our knowledge, no study has yet to identify a naturally occurring CoNS AIP inhibitor of MRSA agr-IV. MRSA agr-IV skin infections are rare but serious. They are most often associated with staphylococcal scalded skin syndrome and the agr-regulated production of exfoliative toxins (Jarraud et al., 2002; Thoendel et al., 2011). Given our in vitro half-maximal inhibitory concentration findings, we determined whether S. warneri AIP-II could be an effective MRSA agr-IV inhibitor in vivo. We found that injection of 50 μg synthetic AIP-II decreased MRSA agr-IV dermonecrotic lesion size (Figure 2g) throughout the course of infection and protected animals from systemic weight loss (Figure 2h) and skin injury (Figure 2i).
Together, our results suggest an important protective role for S. warneri quorum-sensing crosstalk on the skin. More work is needed to determine the distribution of these agr types on healthy skin and whether the absence of S. warneri may be correlated with disease. Further studies may reveal other mechanisms of S. warneri colonization resistance, such as the production of antimicrobial peptides (Nakatsuji et al., 2017). In future translational applications, S. warneri or its molecular products could potentially be harnessed to treat AD or other skin diseases in a biotherapeutic approach (Nakatsuji et al., 2021).
Supplementary Material
ACKNOWLEDGMENTS
MMS was supported by the National Institute of Allergy and Infectious Diseases Ruth L. Kirschstein National Research Award Predoctoral Fellowship AI157052. ZLB was supported by the National Center for Complementary and Integrative Health Ruth L. Kirschstein National Research Award Training Grant T32AT008938. ARH and NBC were supported by the National Institute of Allergy and Infectious Diseases grant AI153185.
Abbreviations:
- AD
atopic dermatitis
- agr
accessory gene regulator
- AIP
autoinducing peptide
- CM
conditioned media
- CoNS
coagulase-negative staphylococci
- MRSA
methicillin–resistant Staphyloccocus aureus
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2022.05.1092
Data availability statement
All data needed to evaluate the conclusions in the letter are present in the letter and/or the Supplementary Materials.
REFERENCES
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018;16:143–55. [DOI] [PubMed] [Google Scholar]
- Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med 2017;9:eaal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol 2021;147:955–966.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Irvine AD, Foster TJ. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol 2018;26:484–97. [DOI] [PubMed] [Google Scholar]
- Gless BH, Bojer MS, Peng P, Baldry M, Ingmer H, Olsen CA. Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation. Nat Chem 2019;11:463–9. [DOI] [PubMed] [Google Scholar]
- Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun 2002;70:631–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos WE, Schleifer KH. Isolation and characterization of staphylococci from human skin II. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int J Syst Bacteriol 1975;25:62–79. [Google Scholar]
- Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med 2020;12:eaay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 2017;9:eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Cheng JY, Gallo RL. Mechanisms for control of skin immune function by the microbiome. Curr Opin Immunol 2021;72:324–30. [DOI] [PubMed] [Google Scholar]
- Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol 2014;196:3482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M, Echner H, Voelter W, Götz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 2001;69:1957–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlet CP, Brown MM, Horswill AR. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol 2019;27:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the Staphylococci. Chem Rev 2011;111:117–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 2019;11:eaat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the letter are present in the letter and/or the Supplementary Materials.