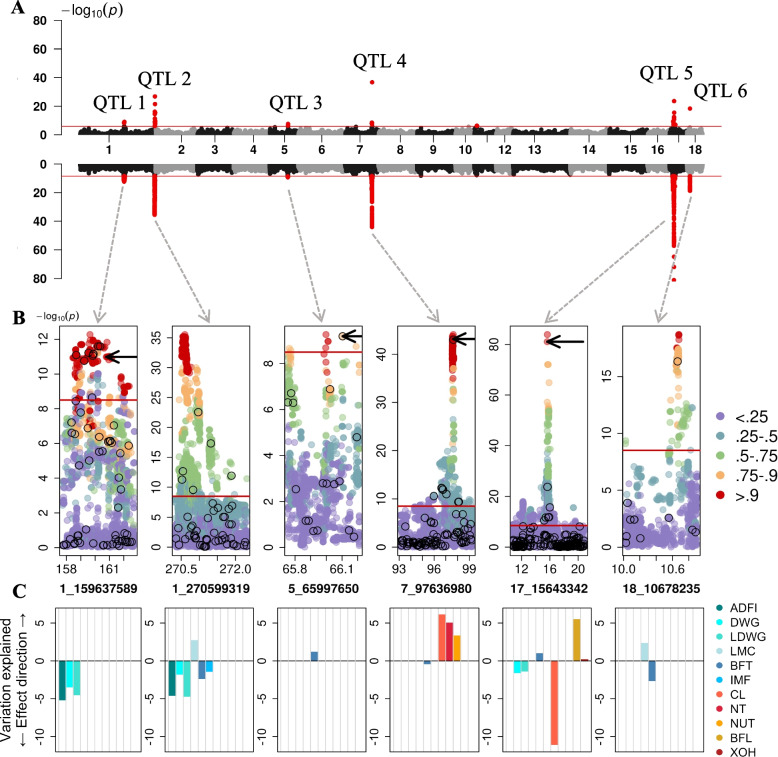
Fig. 2.
Fine mapping of six QTL detected by metaGWAS. A Manhattan plots from array (upper) and imputed sequence (bottom) variants in metaGWAS with 24 traits. B Linkage disequilibrium between the lead SNPs and all other variants. Black circles mark array SNPs, arrows point to previously proposed causal variants. The red line indicates the genome-wide Bonferroni-corrected significance threshold. C Variation explained (in % of the drEBV variance) by alternative alleles of the lead SNPs in the single traits. Production traits are in blue scale (ADFI—Average daily feed intake; DWG—Daily weight gain on test; LDWG—Lifetime daily weight gain; LMC—Lean meat content; BFT—Back fat thickness; IMF—Intramuscular fat content in loin), and conformation traits in red scale (CL—Carcass length; NT—Number of teats—both sides; NUT—Number of underdeveloped teats; BFL—Bent to pre-bent, front legs; XOH—X- to O-legged)