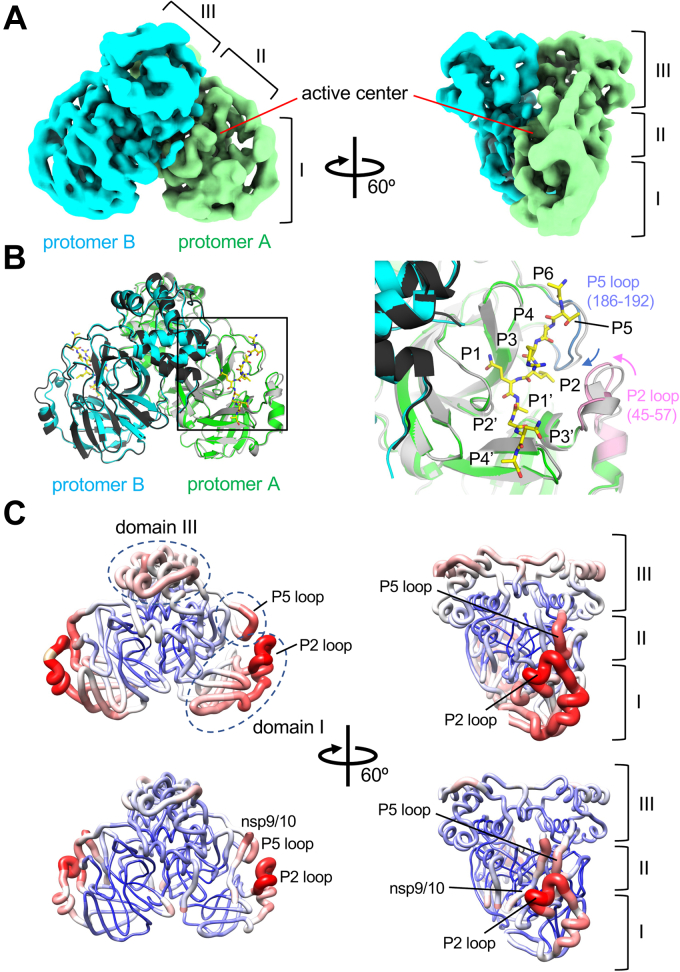
Figure 3.
Cryo-EM structure of wild-type Mpro.A, cryo-EM density map of Mpro. Density maps corresponding to each protomer of the Mpro dimer are colored and indicated. Three domains and the active center of Mpro are indicated. B, left, comparison of the Mpro structures in the apo form (gray and block) and in the nsp7-10 complex (green, cyan, and yellow); Right, a magnified view of the active center of the Mpro (boxed area of protomer A in the right panel). The flexible P2 (pink) and P5 loops (blue) around the substrate binding cleft move toward the nsp9/10 recognition site in the Mpro and polyprotein complex (indicated by pink and blue arrows). C, comparison of the B-factor distributions in the apo form (top) and in the nsp7-10 complex (bottom) of Mpro. Cartoon representation of the models with gradients of color (blue, white to red) and thickness (narrow to wide) reflecting the scale of the B factors (low to high). Domains and loops of the apo-form Mpro showing a higher B-factor compared with the Mpro and nsp7/10 complex are indicated by black dashed ovals. Mpro, main protease.