ED Fig 1. Dataset description and representative driver gene maps.
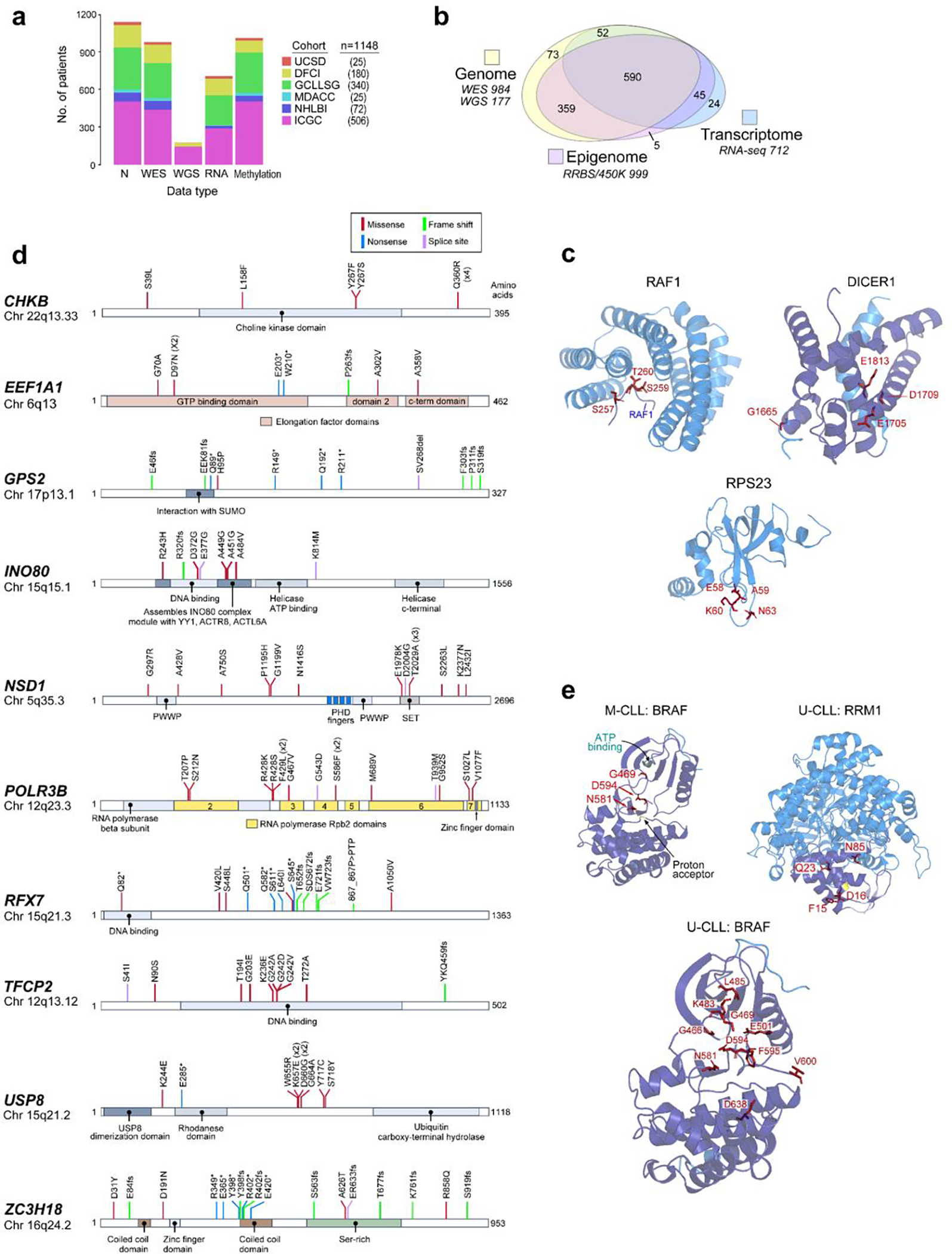
a. Full dataset (n=1148), with contributions by cohort and data type delineated (see Supplementary Table 1). b. Numbers of samples with genomic, epigenomic, and transcriptomic data. c. 3D protein structures of representative genes identified by CLUMPS in pan-CLL analysis (n=984, see Supplementary Table 5). Mutated residues - red labels. A peptide from RAF1 (designated at bottom-center, in complex with 14–3-3 zeta) shows clustered mutations around S259, whose phosphorylation regulates RAF1 activity and is a cancer mutational hotspot90 that, when mutated, perturbs the interaction with the 14–3-3 zeta and upregulates RAF1 kinase activity91,92. In DICER1, mutations occur in the RNase III domain (purple), including the cancer hotspot residue E181321,24. This region is critical for Mg2+ binding and is required for ribonuclease activity to process microRNAs and mediate post-transcriptional gene regulation93. RPS23 mutations are clustered in a conserved loop of the ribosomal decoding center, surrounding P62, whose post-translational hydroxylation affects translation termination accuracy94. These RPS23 mutations have a median CCF >80% (Extended Data Fig. 6d; Supplementary Table 3). d. Individual mutations maps of selected novel, putative driver genes. Mutation subtype and position are shown. e. Selected genes identified by CLUMPS in IGHV subtypes; mutated residues - red. Although BRAF was not identified as a potential M-CLL driver via MutSig2CV (see Extended Data Fig. 3, Methods), CLUMPS revealed three mutated sites clustered in the kinase domain (purple) that are cancer hotspots24, thus confirming BRAF as a shared driver (left). Mutated residues in BRAF in U-CLL (bottom) are shown for comparison, revealing a greater number of clustered mutations relative to M-CLL. In U-CLL, novel mutations were found in RRM1 (right). Somatic alterations were clustered in the N-terminal ATP-binding site (purple) and therefore have potential to impact enzymatic activity95.
