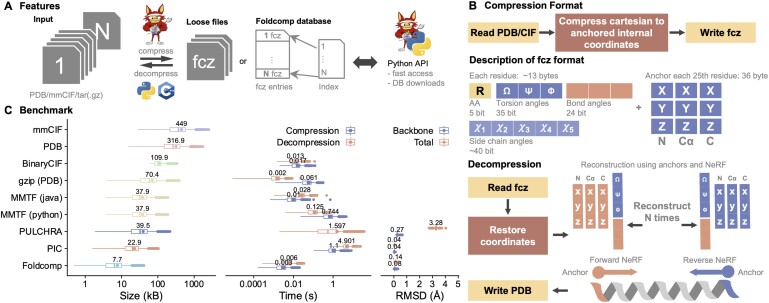
Figure 1.
(A) Foldcomp is a library to compress, store and index protein structures. Foldcomp is written in C++ and comes with a command line and Python interface to compress, decompress, and access structures. (B) Compression takes 3D atom coordinates stored in PDB/mmCIF format as input and calculates and stores all internal coordinates, backbone torsions, and bond angles, and additionally, for every Nth residue (by default every 25th) the 3D atom coordinates for the N, C, and C-alpha atoms as anchor coordinates in its fcz format. By using anchors, we can prevent the accumulation of decoding errors. Decompression uses the anchor coordinates and internal coordinates to reconstruct the 3D atom coordinates by first extending the coordinates from N-terminal to C-terminal (forward) and then from C-terminal to N-terminal (backward) using Natural Extension Reference Frames (NeRF; Parsons et al. 2005), followed by averaging the coordinates in between. Averaging reduces the reconstruction error by approximately a factor of two. (C) Comparison of file size (left), compression/decompression speed (middle), and backbone/all-atoms reconstruction error (right) for lossless and lossy protein structure compressors using the Saccharomyces cerevisiae proteome from the AlphaFold DB