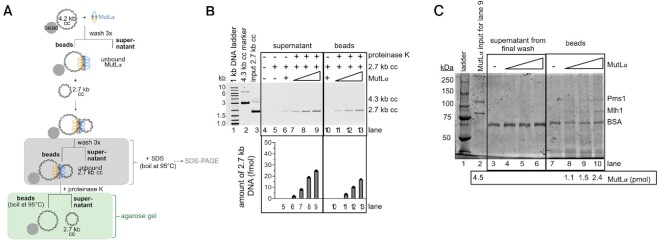
Figure 2.
An excess of MutLα is required to promote DNA–DNA associations. (A) Schematic depicting DNA pull-down experiment. Biotinylated 4.3 kb covalently closed (cc) plasmid is immobilized on streptavidin beads. MutLα is bound to immobilized DNA and a 2.7 kb unbiotinylated plasmid is added to determine if the MutLα that is already bound to DNA can pull-down additional DNA molecules. Fractions that are analyzed by agarose gel are shown in green and fractions that are denatured and analyzed by SDS-PAGE are shown in grey. (B) Agarose gel assaying supernatant and bead-bound fractions. Lane 1 contains 1 kb plus DNA ladder (NEB), lane 2 contains a 4.3 kb DNA marker and lane contains DNA inputs for 2.7 kb (135 ng, 77 fmol) plasmids. In lanes 6 and 10, 8.0 pmol MutLα was added. In lanes 7–9 and 11–13, MutLα titration amounts are 2.6 pmol, 8 pmol, and 13.3 pmol respectively. Amount of recovered 2.7 kb DNA (fmol) relative to the input is shown in bar graph below the gel. Number of replicates is 2. (C) SDS-PAGE analysis of supernatant and bead-bound fractions. Lane 1 contains Precision Plus protein ladder (BioRad). Lane 2 contains the MutLα input for lane 9 (8.0 pmol). The Mlh1 subunit weighs 87 kDa and the Pms1 subunit weighs 99 kDa.