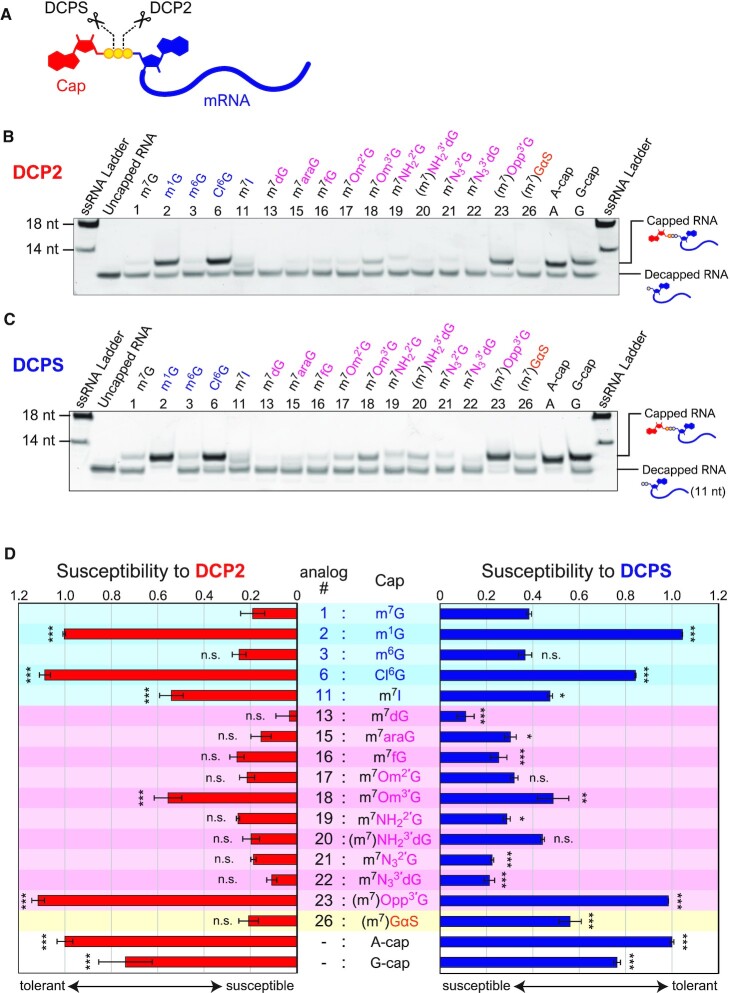
Figure 3.
Susceptibility of cap-modified RNAs to decapping enzymes. (A) Decapping by Decapping enzymes. Cap structure is degraded by DCP2 and DCPS. Each decapping enzyme cleaves 5′-5′ triphosphate at a different position. (B) An example of decapping assay by DCP2. The lower and upper bands indicate the decapped RNA and remaining capped RNA, respectively. (C) An example of decapping assay by DCPS. (D) The sensitivities to decapping enzymes are shown based on the remaining amount of capped RNA. The values were normalized with the value of A-cap. Statistical significance to m7G-capped RNA (#1): n.s.: not significant, * P < 0.05, ** P < 0.01, *** P < 0.001 (Dunnett's test). m7; expected that the N7 is almost fully methylated; (m7): N7 is partialy methylated.