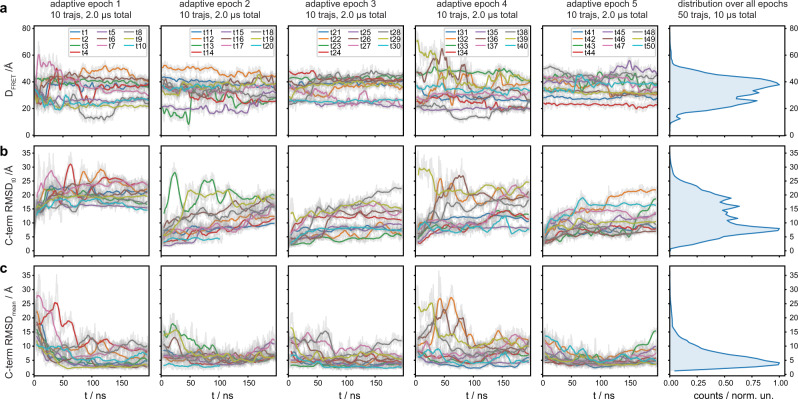
Fig. 6. The flexibility of the β2AR CT observed in MD simulations.
a–c The flexibility of the β2AR CT observed in MD simulations and the related histogram of all the values of the CT fluctuations around each trajectory’s average structures. The first five columns represent the five adaptive sampling epochs, each epoch consisting of ten individual, unbiased, 200 ns long MD trajectories of the β2AR. Each row shows the time traces for different geometrical parameters. a Minimum distance between CYS148 and CYS378 heavy-atoms, tracking the distance between dyes in the smFRET experiments. This parameter was used as the “exploit” component in the adaptive sampling. We observe a high range of distances sampled in all 50 trajectories, indicating a high structural flexibility of CT in the nanosecond timescale: Only in a few trajectories does the CYS-CYS distance remain constant (e.g. the red curve in the last epoch, trajectory 44). In most cases, it changes by tens of Angstrom (e.g. the green curve of the second epoch, trajectory 13). b Root-mean-square-deviation (RMSD) of the CT backbone atoms, monitoring the structural changes of the CT with respect to each trajectory’s starting frame. Strikingly, The CT moves away quickly from the modeled starting conformation by up to 30 Å in the first epoch. The disordered nature of CT is reflected by the high variability of different conformations sampled in all epochs. c RMSD of the CT backbone atoms, monitoring the fluctuations of the CT around the trajectory’s average structure. The wide range of values hints at a rather flat free-energy landscape underlying the dynamics, which is characteristic of structurally disordered proteins. Time-traces show the running-averages as solid lines (smoothing window of 50 ns) overlaid on top of the raw time-trace, shown in gray in the background. The distributions on the rightermost panels are computed using the raw data.