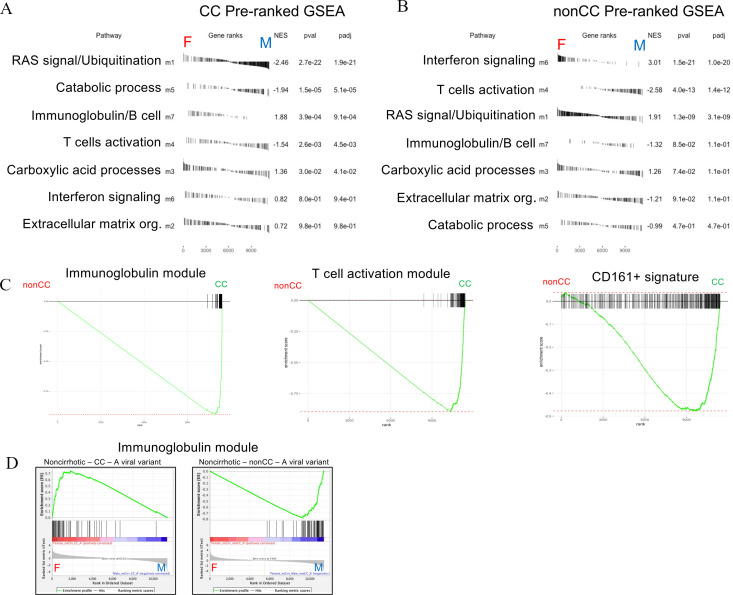
Figure 6.
Gene module enrichment between sexes in CC and non-CC genotypes. Preranked (after covariate adjustments) gene set enrichment analyses (GSEAs) showing sex enrichments of all weighted gene coexpression network analysis (WGCNA) modules (m1–m7) in CC (A) and non-CC (B) samples. Module enrichments are ranked by statistical significance (adjusted p values). (C) Preranked GSEAs with enrichment curves for immunoglobulin module (m7, left plot, false discovery rate (FDR) <<0.01), T-cell module (m4, middle plot, FDR<<0.01) and Mucosal Associated Invariant T (MAIT) cell signature (CD161+, right plot, FDR=0.003) between IFNL4 (interferon lambda 4) genotypes (non-CC versus CC) in overall analysis (all covariates control). Standard GSEAs (D) showing sex enrichments of immunoglobulin module in subsets of non-cirrhotic-CC with A viral variant in pos2570 (CC-A) (left plot, FDR<<0.01) and non-cirrhotic-non-CC with same viral variant (non-CC-A, FDR<<0.01) (right plot).