Abstract
Whole slide imaging (WSI) allows pathologists to view virtual versions of slides on computer monitors. With increasing adoption of digital pathology, laboratories have begun to validate their WSI systems for diagnostic purposes according to reference guidelines. Among these the College of American Pathologists (CAP) guideline includes three strong recommendations (SRs) and nine good practice statements (GPSs). To date, the application of WSI to cytopathology has been beyond the scope of the CAP guideline due to limited evidence. Herein we systematically reviewed the published literature on WSI validation studies in cytology. A systematic search was carried out in PubMed‐MEDLINE and Embase databases up to November 2021 to identify all publications regarding validation of WSI in cytology. Each article was reviewed to determine if SRs and/or GPSs recommended by the CAP guideline were adequately satisfied. Of 3963 retrieved articles, 25 were included. Only 4/25 studies (16%) satisfied all three SRs, with only one publication (1/25, 4%) fulfilling all three SRs and nine GPSs. Lack of a suitable validation dataset was the main missing SR (16/25, 64%) and less than a third of the studies reported intra‐observer variability data (7/25, 28%). Whilst the CAP guideline for WSI validation in clinical practice helped the widespread adoption of digital pathology, more evidence is required to routinely employ WSI for diagnostic purposes in cytopathology practice. More dedicated validation studies satisfying all SRs and/or GPSs recommended by the CAP are needed to help expedite the use of WSI for primary diagnosis in cytopathology.
Keywords: CAP guideline, cytopathology, digital pathology, systematic review, validation, WSI
To date, the application of WSI to cytopathology has been beyond the scope of the CAP guideline, due to limited evidence. Whilst the CAP guideline for WSI validation in clinical practice helped the widespread adoption of digital pathology, more evidence is required to routinely employ WSI for diagnostic purposes in cytopathology practice.

1. INTRODUCTION
Digital pathology consists of viewing, sharing, and/or analysing digitised pathology glass slides employing computer‐based technology. 1 There are numerous clinical (e.g., primary diagnosis, telepathology, image analysis) and non‐clinical (e.g., research, education) applications of digital pathology. Imaging technology related to digital pathology has evolved over time, from static images (microphotographs of a field of view on a slide) to dynamic images (transmission of images in real time), and more recently to whole slide imaging (WSI). WSI technology refers to scanning glass slides to generate digital slides that can be viewed on a computer monitor to recreate a virtual experience that is similar to examining the glass slides with a traditional light microscope. 2
To demonstrate that this technology works safely for diagnostic patient care and that it can accordingly be adopted for routine clinical work, WSI systems should ideally undergo validation before deployment in clinical service. The crux of such a validation study is to ensure that pathologists' diagnoses using WSI are as accurate as those rendered with glass slides and a light microscope. To assist pathology laboratories with this validation process, in 2013 the College of American Pathologists (CAP) published a specific guideline on how to validate WSI for diagnostic purposes. The CAP guideline incorporated 12 statements to guide pathology laboratories. 3 The CAP guideline was subsequently updated in 2021, 4 and differed from the previous publication because a Grading of Recommendation Assessment, Development, and Evaluation (GRADE) 5 framework was adopted to evaluate available evidence. Moreover, the concept of good practice statements (GPS) was introduced. GPSs differ from strong recommendations (SRs) because while they support important issues they lack the published evidence typically needed for a recommendation. The updated CAP guideline comprising three SRs and nine GPSs is summarised in Table 1.
TABLE 1.
Strong recommendations (SRs) and good practice statements (GPSs) from the 2021 College of American Pathologists guideline for validation of whole slide imaging systems
| Item | Description |
|---|---|
| SR 1 | The validation process should include a sample set of at least 60 cases for one application, or use case (e.g., haematoxylin–eosin–stained sections of fixed tissue, frozen sections, haematology), that reflect the spectrum and complexity of specimen types and diagnoses likely to be encountered during routine practice. The validation should include another 20 cases to cover additional applications such as immunohistochemistry or other special stains if these applications are relevant to an intended use and were not included in the 60 cases mentioned above. |
| SR 2 | The validation study should establish diagnostic concordance between digital and glass slides for the same observer (i.e., intra‐observer variability). If concordance is less than 95%, laboratories should investigate and attempt to remedy the cause. |
| SR 3 | A washout period of at least 2 weeks should occur between viewing digital and glass slides |
| GPS 1 | All pathology laboratories implementing WSI technology for clinical diagnostic purposes should carry out their own validation studies. |
| GPS 2 | Validation should be appropriate for and applicable to the intended clinical use and clinical setting of the application in which WSI will be used. Validation of WSI systems should involve specimen preparation types relevant to intended use (e.g., formalin‐fixed, paraffin‐embedded tissue; frozen tissue; immunohistochemical stains). If a new application for WSI is contemplated, and it differs materially from the previously validated use, a separate validation for the new application should be performed. |
| GPS 3 | The validation study should closely emulate the real‐world clinical environment in which the technology will be used. |
| GPS 4 | The validation study should encompass the entire WSI system. It is not necessary to separately validate each individual component (eg, computer hardware, monitor, network, scanner) of the system or the individual steps of the digital imaging process. |
| GPS 5 | Laboratories should have procedures in place to address changes to the WSI system that could impact clinical results. |
| GPS 6 | Pathologists adequately trained to use the WSI system must be involved in the validation process. |
| GPS 7 | The validation process should confirm all of the material present on a glass slide to be scanned is included in the digital image. |
| GPS 8 | Documentation should be maintained recording the method, measurements, and final approval of validation for the WSI system to be used in the anatomic pathology laboratory. |
| GPS 9 | Pathologists should review cases/slides in a validation set in random order. This applies to both the review modality (ie, glass slides or digital) and the order in which slides/cases are reviewed within each modality. |
Most validation guidelines related to WSI for diagnostic use, including the aforementioned published CAP recommendations, do not specifically include cytology. In fact, the authors of the CAP guideline underline that at the time of publication, due to lack of published evidence, validation of WSI in cytology was considered beyond the scope. Indeed, the adoption of digital cytology has lagged behind that of digital histopathology for several reasons, such as the difficulty of scanning cytology material on glass slides in different focal planes using Z‐stacking. 6 Not surprisingly, published clinical validation studies in cytology are less numerous than those involving surgical pathology.
The aim of this study was accordingly to investigate the published literature concerning the validation of WSI systems specifically in cytology, with reference to the CAP guideline.
2. MATERIALS AND METHODS
2.1. Literature search and article screening
The review question was formulated according to a Population, Index, Comparator, Outcome (PICO) model. Population was represented by a series of cytology cases collected retrospectively or prospectively for the validation study; the Index was the WSI modality for pathology cases, while the Comparator was represented by conventional light microscopy. Outcome was represented by concordance between a diagnosis rendered with WSI and light microscopy, the latter being taken as the reference standard. The main aim of the study was to investigate the adherence of validation studies for WSI in cytology to the CAP guideline. Studies represented by abstract only with limited information were excluded.
A systematic review was conducted according to standard methods and reporting in accordance with the Preferred Reporting Items for Systematic reviews and Meta‐Analysis (PRISMA). 7 The databases PubMed and Embase were systematically searched up to 20 November 2021 to identify any article regarding a validation study of WSI in cytology. The search strategy comprised combinations of the terms “digital pathology,” “validation,” and “cytology” with their conceptual aliases and variations, adequately adapted to the two databases' search engines. Four authors (AE, IG, NS, PA) independently reviewed all article titles and abstracts with the aid of the Rayyan reference manager web application. 8
Papers dealing with digital pathology other than human cytology (e.g., histopathology, frozen sections of surgical specimens, etc.), with static and dynamic images, or with animal or experimental models, were excluded, as well as papers in languages other than English. Full texts of the articles fulfilling initial screening criteria were acquired and reviewed against the eligibility criteria. Any disagreement with respect to inclusion of a particular article was resolved by consensus.
2.2. Data extraction
Two investigators (SN, PA) independently extracted data from the included studies with a standardised form. Data extracted included: author(s) and publication year, country of origin for the research, total number of cytological cases, site(s) of origin of the cytological material, and compliance with the CAP guideline criteria for SRs and GPSs.
3. RESULTS
3.1. Overview of the papers
A flow diagram of the screening, selection, and exclusion of articles for this review is shown in Figure 1. Briefly, 3963 papers were found and screened with the aid of the Rayyan reference manager web application. 8 After title and abstract screening were undertaken, 69 papers were selected as potentially relevant to the review and after subsequent full text assessment 44 articles were then excluded. Thus, overall 25 papers were included in our review, representing studies published between 2001 and 2021. A cumulative total of 1994 cytological cases were included (ranging from 5 to 505 cases per study), and comprised case series from Australia, 9 Canada, 10 China, 11 Colombia, 12 India, 13 Italy, 14 Japan, 15 the Netherlands, 16 Norway, 17 , 18 Poland, 19 Portugal, 20 , 21 Taiwan, 22 the UK, 23 and the USA. 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 . Of the 25 papers included, eight (32%) dealt with gynaecological cytology, 14 , 16 , 23 , 24 , 25 , 27 , 32 , 33 and the remainder (68%) with non‐gynaecological cytology including three with thyroid cytology, 20 , 21 , 30 one with thoracic cytology, 19 one with central nervous system cytology, 10 one with breast cytology, 15 one with peripheral blood smears, 28 and ten with specimens derived from different anatomic sites. 9 , 11 , 12 , 13 , 17 , 18 , 22 , 26 , 29 , 30 , 31
FIGURE 1.
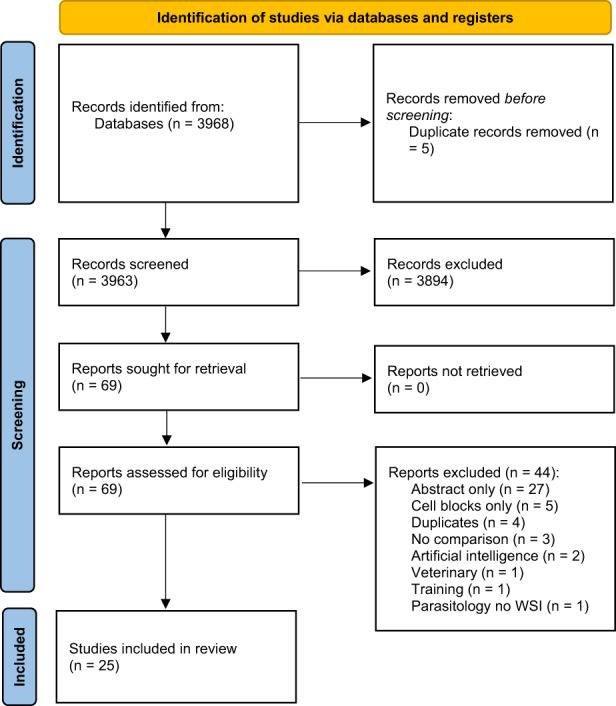
Search flow diagram, adapted from the PRISMA flow diagram template (Page et al 7 ). PRISMA, Preferred Reporting Items for Systematic reviews and Meta‐Analysis
3.2. Assessment of strong recommendations (SRs) and good practice statements (GPSs)
Only 4/25 papers (16%) satisfied the criteria for all three SRs, 13 , 17 , 28 , 32 and only 1/25 paper (4%) satisfied all three SRs and nine GPSs. 13 Moreover, 10/25 papers (40%) did not indicate compliance for even a single SR, while 24/25 (96%) failed to demonstrate compliance for at least one GPS. 9/25 (36%) papers addressed a single SR only, while 16/25(64%) did not address any SR, making SRs the most ignored parameters. On the other hand, 25/25 studies (100%) satisfied GPS 1, 2, and 4. An overall depiction of the included studies and their compliance with specific SRs and GPSs is provided in Table 2.
TABLE 2.
Characteristics of the included studies and their compliance with each SR and GPS from the College of American Pathologists guideline for validation of whole slide imaging systems
| Authors | Year | Country | Case number | Anatomic site | Stain | Digital system | SR 1 | SR 2 | SR 3 | GPS 1 | GPS 2 | GPS 3 | GPS 4 | GPS 5 | GPS 6 | GPS 7 | GPS 8 | GPS 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Steinberg 24 | 2001 | USA | 10 | Cervicovaginal | Papanicolaou |
BLISS™ system; WebSlide Browser™ |
||||||||||||
| Marchewsky 23 | 2006 | USA | 20 | Cervicovaginal | Papanicolaou |
ScanScope (Aperio); VG700b displays |
||||||||||||
| Dee 25 | 2007 | USA | 5 | Cervicovaginal | Not specified | MicroBrightField software | ||||||||||||
| Slodkowska 19 | 2009 | Poland | 28 | Thoracic | Not specified | Coolscope (Nikon); ScanScope (Aperio) | ||||||||||||
| Evered 33 | 2010 | UK | 20 | Cervicovaginal | Not specified | NanoZoomer HT (Hamamatsu Photonic KK) | ||||||||||||
| Gould 10 | 2012 | Canada | 30 | Brain | Not specified |
NanoZoomer HT 2.0 (Hamamatsu Photonic KK); NDP.view software; HP L2245 wg monitor |
||||||||||||
| House 26 | 2013 | USA | 21 | Mixed | Papanicolaou, Diff‐Quik, H&E, Gomori Methenamine Silver |
ScanScope XT (Aperio); Spectrum and ImageScope software (Aperio) |
||||||||||||
| Wright 27 | 2013 | USA | 11 | Cervicovaginal | Papanicolaou | BioImagene iScan Coreo Au 3.0 (Ventana) | ||||||||||||
| Donnelly 32 | 2013 | USA | 192 | Cervicovaginal | Not specified |
BioImagene iScan Coreo Au (Ventana); ImageViewer 3.0.0.0 software (Ventana) |
||||||||||||
| Gerhard 20 | 2013 | Portugal | 222 | Thyroid | Diff Quik | NanoZoomer HT 2.0 (Hamamatsu) | ||||||||||||
| Gomez‐Gelvezet 28 | 2015 | USA | 100 | Peripheral blood smears | Wright‐Giemsa |
ScanScope (Aperio); ImageScope software (Aperio) |
||||||||||||
| Vodovnik 18 | 2015 | Norway | 36 | Mixed | Romanowsky or Romanowsky/Papanicolaou |
ScanScope AT turbo (Leica); ImageScope software (Leica); SymPathy LIMS (Tieto) |
||||||||||||
| Hang 22 | 2015 | Taiwan | 10 | Mixed | Papanicolaou |
SCN400 (Leica); Aperio eSlide Manager (Leica) |
||||||||||||
| Mukherjee 30 | 2015 | USA | 12 | Thyroid | Papanicolaou |
iScan Coreo Au (Ventana); ImageViewer 3.1 software (Ventana) |
||||||||||||
| Arnold 31 | 2015 | USA | 21 | Paediatric | Not specified |
Model XT (Aperio); ImageScope software (Leica) |
||||||||||||
| Hanna 29 | 2017 | USA | 30 | Mixed | Papanicolaou, Diff‐Quik, H&E |
ScanScope XT (Leica); ImageScope software (Leica) |
||||||||||||
| Vodovnik 17 | 2018 | Norway | 204 | Mixed | Romanowsky |
ScanScope AT Turbo (Aperio); ImageScope software (Aperio); SymPathy LS (Tieto) |
||||||||||||
| Bongaerts 16 | 2018 | Netherlands | 505 | Cervix | Not specified |
250 Flash II scanner (3D Histech); Pannoramic viewer software (3D Histech) |
||||||||||||
| Huang 11 | 2018 | China | 100 | Mixed | Not specified |
200 Android smartphones; Aperio AT2 |
||||||||||||
| Ross 9 | 2018 | Australia | 56 | Mixed | Papanicolaou, Romanowsky |
Aperio ScanScope XT (Leica); ImageScope software (Aperio); Collibio web links (Pixcelldata) |
||||||||||||
| Yamashiro 15 | 2019 | Japan | 23 | Breast | Not specified | Panoptiq software | ||||||||||||
| Mosquera‐Zamudio 12 | 2019 | Colombia | 17 | Mixed | Papanicolaou |
NanoZoomer HT 2.0 (Hamamatsu Photonics); Panoptiq system |
||||||||||||
| Canberk 21 | 2020 | Portugal | 227 | Thyroid | Papanicolaou, Diff‐Quik |
P250 Flash III; PMidi; PDesk (3DHistech) |
||||||||||||
| Rajaganesanet 13 | 2021 | India | 60 | Mixed | Papanicolaou, May–Grunwald–Giemsa | Not specified | ||||||||||||
| Negri 14 | 2021 | Italy | 34 | Cervix | Papanicolaou |
D‐Sight scanner (Menarini); JVSview software |
Note: Shadings indicate whether the study satisfied (green), did not satisfy (red), or did not report data (yellow) for each SR or GPS. Abbreviations: SR, strong recommendation; GPS, good practice statement.
4. DISCUSSION
Whole slide imaging technology involves the acquisition of digital images of entire pathology glass slides. 34 WSI has numerous benefits such as portability of pathologists, easy sharing of digital slides, side‐by‐side comparison of slides on a monitor, image analysis, and several other useful applications. 35 As a result, WSI has gained popularity for clinical purposes such as teleconsultation, as well as educational purposes and research activity. 36 Systematic reviews on the concordance of WSI versus viewing glass slides using light microscopy have demonstrated that the overall diagnostic concordance between these two modalities is greater than 90%, sometimes with an excellent κ coefficient. 37 , 38 However, the application of WSI for cytology has been problematic due to several technical reasons (e.g., cytology smears may cover the entire glass slide surface, cytology material has areas of variable thickness, there may be obscuring material, and cell clusters in three‐dimensions make it difficult to focus in just one plane). 39 For these reasons, publications regarding WSI in cytology are limited and concordance results with glass slides reported for digital cytology versus histology differ. 39 Nevertheless, this gap is closing as more publications provide data supporting the diagnostic use of WSI for cytopathology. 40 , 41
4.1. Strong recommendations (SRs)
SR 1 states that the validation process should include a sample set of at least 60 cases for one application or use case that reflect the spectrum and complexity of specimen types and diagnoses likely to be encountered during routine practice. The validation process should include another 20 cases to cover additional applications such as immunohistochemistry or other special stains if these applications are relevant. However, the Royal College of Pathologists best practice recommendations for implementing digital pathology in 2018 stated that the sample size and duration of the validation process can vary according to specific circumstances. 42 This is true particularly for studies that focus on rare pathologies for which it may prove difficult to recruit enough cases to meet SR 1.
Our systematic review indicates that 68% of included studies did not meet the required number of 60 cases, with 24% of the studies reporting less than 20 cases, and with a minimum of only five cases in one study. 25 In our opinion, sample size is an important criterion and we accordingly recommend this be adhered to in future validation studies of WSI in cytology. As explained by the CAP in their updated guideline, a reasonable number of cases will be needed in the validation process in order to include enough cases that represent the entire spectrum and proportion of diagnoses likely to be encountered in a particular clinical setting.
SR 2 states that validation studies should establish diagnostic concordance between digital and glass slides for the same observer. The original diagnoses for selected cases to be used may have been made by pathologists other than those completing the validation, thereby providing additional usable information on inter‐observer variability. The recommendation from the CAP is that, although all discordances between WSI and glass slide diagnoses discovered during the validation need to be reconciled, laboratories should only be concerned if their overall WSI‐to‐glass slide concordance is less than 95%. More than half (16/25, 64%) of the studies in our review indicated intra‐observer concordance below this standard. This may be partially explained by the fact that most of the papers were conducted in a setting where the diagnosis with light microscopy and diagnosis with digital slides were rendered in different places by different pathologists. In 2015 Vodovnik et al 18 compared the timing of digital and microscopic diagnosis in routine practice, finding that digital cases were diagnosed more quickly. However, no quantitative data about concordance between digital and light microscopy was assessed.
Intra‐observer variability was only established in 28% of the papers. Donnelly et al 32 evaluated 192 gynaecological cases, and reported an overall intra‐rater concordance for each of the five investigators in their study, ranging from 89% to 97%, with only one meeting the 95% criterion recommended by the CAP. Moreover, in the work by Gerhard et al 20 intra‐observer variability was only reported for one of the two physicians, with an intra‐observer concordance of 77.5% involving 222 thyroid cases. Gomez‐Gelvez et al 28 assessed 100 peripheral blood smears and reported an intra‐observer variability for each of four participants, with concordance rates ranging from 88% to 94%. Discordances reported that did not impact patient management (defined as minor discordances) were 8%, 8%, 4%, and 4% for the separate evaluators; conversely, major discordances potentially affecting patient management were 4%, 2%, 2%, and 4%, respectively, for each reader.
A more accurate intra‐observer concordance evaluation in terms of Cohen's Kappa coefficient was reported in the validation study by Rajaganesan et al 13 that included 60 cytology cases, with two pathologists who reported almost perfect concordance (k = 0.8), another two pathologists who had substantial intra‐observer agreement (k = 0.6‐0.8), and another one that had moderate concordance (k = 0.4‐0.6). Diagnostic concordance in the study by Hanna et al 29 was assessed on 30 cases, including five cell blocks, and compared the results for two digital systems (Panoptiq and an Aperio system). Indeed, as recommend by the CAP, each scanner requires its own set of validation cases. Furthermore, the study by Vodovnik et al 17 that included 600 total cases, 204 of which were cytology specimens, did not report on concordance rates. Only six of their cases revealed minor discordances, none of which involved the cytopathology cases. Hence, supposedly the intra‐observer concordance for their cytology cases was 100%. Mukherjee et al 30 reported an intra‐observer concordance for 12 thyroid cases that were scanned with three, five, and seven focal planes. Their intra‐observer concordance ranged from 92% to 100%.
Finally, SR 3 states that a washout period of at least 2 weeks should occur between viewing digital and glass slides. This recommendation is intended to address the issue of recall bias when cases are reviewed using different modalities by the same observer. 43 A significant proportion (44%) of the studies included in our review met this criterion, as all studies reporting intra‐observer variability respected at least a two‐week washout interval, apart from the publication by Mukherjee et al 30 where the washout period was 2 days.
4.2. Good practice statements (GPSs)
In the CAP guideline, GRADE introduced the concept of GPS for several issues where published evidence was lacking to support specific recommendations. Overall, 50% of all the GPSs were met, compliance was not specified for 47% of GPSs, and 3% were not satisfied at all. Evaluation of included publications for fulfilment of GPSs was difficult given that extensive descriptions of the study setting were not always available. GPS 1, 2, and 4 were satisfied in all 25 studies. GPS 3 states that the validation study should closely emulate the real‐world clinical environment in which the technology will be used and laboratories are free to incorporate whatever they feel would be appropriate to achieve this goal. In our review, while six studies failed to report, only two studies provided data that did not comply with this parameter. Namely, Bongaerts et al, 16 while evaluating WSI for cervical cytology, enriched their samples with high‐grade squamous intraepithelial lesion (HSIL) cases, thereby increasing the vigilance to identify such lesions. Also, Dee et al 25 evaluated the effectiveness of 3‐D versus 2‐D virtual microscopy as adjuncts to education and assessment in cervical cytology. Although 3‐D virtual microscopy systems were on the market or under development at the time of the study, they were not yet were fully integrated for a rapid pan and view with instantaneous focusing capability. Results reported a general consensus that virtual cervical cytology slides would be a useful augmentation to education and testing; however, there was minimal enthusiasm for using virtual slides to replace glass slides. GPS 5 states that laboratories should have procedures in place to address changes to the WSI system that could impact clinical results. Only two studies fulfilled this criterion.
GPS 6 states that pathologists adequately trained to use a WSI system must be involved in the validation process. As clearly reported by the CAP, this was not an evidence‐based recommendation. Moreover, no metrics were suggested to determine technical competency of pathologists using WSI systems. Instead, adequate training is best defined at the discretion of the laboratory medical director. 4 The same applies for the number of pathologists participating in the validation process. In our review, for 64% of the studies pathologist training and competency was not specified. Of interest, Hang et al 22 found that participants from educational programs could make diagnostic interpretations using WSI even without prior experience. Similarly, Rajaganesan et al 13 showed that pathologists can adapt to new technologies irrespective of the system used. Similarly, House et al 26 had cytotechnologists with and without digital experience participate in their validation study. On the other hand, in the work by Dee et al, 25 28 out of 79 evaluators were students, without any routine diagnostic experience.
GPS 7 states that the validation process should confirm all the material present on a glass slide is eventually included in the digital image. The CAP guideline highlights the possibility for scans to be missing some or all of the material present on a glass slide, which may have serious clinical and legal consequences. Possible solutions for this issue include digitising all tissue blocks for comparison, viewing thumbnails of entire scanned slides prior to sign out, introducing a quality control step for a technician to check all scanned slides to verify that all material was completely scanned, or using image analysis software to detect missing tissue on virtual slides. In our review, 7/25 studies (28%) met this criterion, 3/25 (12%) did not, and 15/25 (60%) did not specify their compliance. For example, Steinberg et al 24 specified that only 20%‐30% of the cellular area of each slide was digitised. Gomez‐Gelvez et al 28 specified that they did not fulfil GPS 7 due to impractically large‐size files which were too hard to search for scan failures. Similarly, Mukherjee et al 30 scanned less than 40% of each slide, especially if the smear covered more than 75% of the slide surface, to reduce scan time and file sizes. Wright et al 27 reported that in their study the entire area occupied in SurePath slides was scanned while the edges of ThinPrep slides were not scanned.
GPS 8 states that documentation should be maintained recording the method, measurements, and final approval of validation for the WSI system to be used in the anatomic pathology laboratory. Most of the studies, 24/25 (96%), did not specify their compliance with this recommendation. Lastly, GPS 9 states that pathologists should review cases/slides in random order for validation purposes. With regard to our review, only 3/25 studies (12%) met this criterion. However, it should be mentioned that the CAP publication states there is no specific evidence that changing the order in which cases/slides are reviewed actually influences the data collected, so that the relative weight of this GPS on the validation process is in question.
4.3. Final considerations and limitations of the study
This review aimed to highlight the strengths and limitations of various validation studies specific to WSI diagnostic use in cytopathology according to the SRs and GPSs of the CAP guideline. The strength of our systematic review was the inclusion of validation studies conducted according to the CAP guideline. For the SRs, more than half of the included studies did not meet recommendations concerning at least 60 cases to be utilised, reporting adequate concordance measures, and using the recommended two‐week washout period to read cases on different modalities. For the GPSs, adherence in included studies was variable. Of note, the CAP guideline is only a recommendation and thus is not mandatory for all cytology laboratories to follow. A limitation of our review is the small number of studies included. Also, the reason for missing data was not apparent in all of the articles reviewed. Another limitation was the difficulty we had with homogenously evaluating SRs and GPSs. SRs are numerical parameters, which can be evaluated objectively (i.e., number of cases, washout period of at least 2 weeks), whereas GPSs are more subjective items which may accordingly be interpreted differently by reviewers.
5. CONCLUSION
Increasing global experience and published data support the diagnostic use of WSI for cytopathology. However, extensive validation studies for such diagnostic use in routine cytology practice is still required. Most publications to date about validation studies using WSI for diagnostic use in cytology failed to satisfy many of the recommendations established in the CAP guideline. We accordingly recommend that future validation studies in this field be conducted with more a rigorous study design, in terms of better adherence to the guideline, which will help generate robust evidence to support the successful deployment of WSI for diagnostic use in cytology. Finally, WSI is the first step towards the implementing of artificial intelligence‐aided diagnostics, which may be particularly useful in screening cytology, where most of the cases are negative. This will create the need for new validation criteria which will have to be included in future recommendations and guidelines.
AUTHOR CONTRIBUTIONS
All authors participated in the conception and design or analysis and interpretation of the data. All authors contributed to the drafting of the manuscript and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENT
Open Access Funding provided by Universita degli Studi di Verona within the CRUI‐CARE Agreement.
Antonini P, Santonicco N, Pantanowitz L, et al. Relevance of the College of American Pathologists guideline for validating whole slide imaging for diagnostic purposes to cytopathology. Cytopathology. 2023;34:5‐14. doi: 10.1111/cyt.13178
Pietro Antonini and Nicola Santonicco contributed equally to this study and should be considered joint co‐first authors.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- 1. Pallua JD, Brunner A, Zelger B, Schirmer M, Haybaeck J. The future of pathology is digital. Pathol Res Pract. 2020;216(9):153040. [DOI] [PubMed] [Google Scholar]
- 2. Hanna MG, Parwani A, Sirintrapun SJ. Whole slide imaging: technology and applications. Adv Anat Pathol. 2020;27(4):251‐259. [DOI] [PubMed] [Google Scholar]
- 3. Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137(12):1710‐1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans AJ, Brown RW, Bui MM, et al. Validating whole slide imaging systems for diagnostic purposes in pathology. Arch Pathol Lab Med. 2022;146(4):440‐450. [DOI] [PubMed] [Google Scholar]
- 5. Guyatt G, Oxman AD, Akl EA, et al. GRADE guidelines: 1. Introduction ‐ GRADE evidence profiles and summary of findings tables. J Clin Epidemiol. 2011;64(4):383‐394. [DOI] [PubMed] [Google Scholar]
- 6. Eccher A, Girolami I. Current state of whole slide imaging use in cytopathology: pros and pitfalls. Cytopathology. 2020;31(5):372‐378. [DOI] [PubMed] [Google Scholar]
- 7. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan‐a web and mobile app for systematic reviews. Syst Rev. 2016;5(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ross J, Greaves J, Earls P, Shulruf B, Van Es SL. Digital vs traditional: are diagnostic accuracy rates similar for glass slides vs whole slide images in a non‐gynaecological external quality assurance setting? Cytopathology. 2018;29(4):326‐334. [DOI] [PubMed] [Google Scholar]
- 10. Gould PV, Saikali S. A comparison of digitized frozen section and smear preparations for intraoperative neurotelepathology. Anal Cell Pathol. 2012;35(2):85‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang YN, Peng XC, Ma S, et al. Development of whole slide imaging on smartphones and evaluation with thinprep cytology test samples: follow‐up study. JMIR Mhealth Uhealth. 2018;6(4):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mosquera‐Zamudio A, Hanna MG, Parra‐Medina R, Piedrahita AC, Rodriguez‐Urrego PA, Pantanowitz L. Advantage of Z‐stacking for teleconsultation between the USA and Colombia. Diagn Cytopathol. 2019;47(1):35‐40. [DOI] [PubMed] [Google Scholar]
- 13. Rajaganesan S, Kumar R, Rao V, et al. Comparative assessment of digital pathology systems for primary diagnosis. J Pathol Inform. 2021;9(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Negri G, Macciocu E, Cepurnaite R, et al. Non‐human papilloma virus associated adenocarcinomas of the cervix uteri. Cytologic features and diagnostic agreement using whole slide digital cytology imaging. Diagn Cytopathol. 2021;49(2):316‐321. [DOI] [PubMed] [Google Scholar]
- 15. Yamashiro K, Yoshimi N, Itoh T, et al. A small‐scale experimental study of breast FNA consultation on the internet using Panoptiq. J Am Soc Cytopathol. 2019;8(4):175‐181. [DOI] [PubMed] [Google Scholar]
- 16. Bongaerts O, Clevers C, Debets M, et al. Conventional microscopical versus digital WholeSlide ImagingBased diagnosis of ThinLayer cervical specimens: a validation study. J Pathol Inform. 2018;9(29):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vodovnik A, Aghdam MRF. Complete routine remote digital pathology services. J Pathol Inform. 2018;9(36):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vodovnik A. Diagnostic time in digital pathology: a comparative study on 400 cases. J Pathol Inform. 2016;7(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Słodkowska J, Pankowski J, Siemiatkowska K, Chyczewski L. Use of the virtual slide and the dynamic real‐time telepathology systems for a consultation and the frozen section intra‐operative diagnosis in thoracic/pulmonary pathology. Folia Histochem Cytobiol. 2009;47(4):679‐684. [DOI] [PubMed] [Google Scholar]
- 20. Gerhard R, Teixeira S, Gaspar da Rocha A, Schmitt F. Thyroid fine‐needle aspiration cytology: is there a place to virtual cytology? Diagn Cytopathol. 2013;41(9):793‐798. [DOI] [PubMed] [Google Scholar]
- 21. Canberk S, Behzatoglu K, Caliskan CK, et al. The role of telecytology in the primary diagnosis of thyroid fine‐needle aspiration specimens. Acta Cytol. 2020;64(4):323‐331. [DOI] [PubMed] [Google Scholar]
- 22. Hang JF, Liang WY, Hsu CY, Lai CR. Integrating a web‐based whole‐slide imaging system and online questionnaires in a national cytopathology peer comparison educational program in Taiwan. Acta Cytol. 2015;59(3):278‐283. [DOI] [PubMed] [Google Scholar]
- 23. Marchevsky AM, Khurana R, Thomas P, Scharre K, Farias P, Bose S. The use of virtual microscopy for proficiency testing in gynecologic cytopathology. Arch Pathol Lab Med. 2006;130(3):349‐355. [DOI] [PubMed] [Google Scholar]
- 24. Steinberg DM, Ali SZ. Application of virtual microscopy in clinical cytopathology. Diagn Cytopathol. 2001;25(6):389‐396. [DOI] [PubMed] [Google Scholar]
- 25. Dee FR, Donnelly A, Radio S, Leaven T, Zaleski MS, Kreiter C. Utility of 2‐D and 3‐D virtual microscopy in cervical cytology education and testing. Acta Cytol. 2007;51(4):523‐529. [DOI] [PubMed] [Google Scholar]
- 26. House J, Henderson‐Jackson E, Johnson J, et al. Diagnostic digital cytopathology: are we ready yet? J Pathol Inform. 2013;4(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wright AM, Smith D, Dhurandhar B, et al. Digital slide imaging in cervicovaginal cytology: a pilot study. Arch Pathol Lab Med. 2013;137(5):618‐624. [DOI] [PubMed] [Google Scholar]
- 28. Gomez‐Gelvez JC, Kryvenko ON, Chabot‐Richards DS, Foucar K, Inamdar KV, Karner KH. Comparative analysis reveals potential utility of digital microscopy in the evaluation of peripheral blood smears with some barriers to implementation. Am J Clin Pathol. 2015;144(1):68‐77. [DOI] [PubMed] [Google Scholar]
- 29. Hanna MG, Monaco SE, Cuda J, Xing J, Ahmed I, Pantanowitz L. Comparison of glass slides and various digital‐slide modalities for cytopathology screening and interpretation. Cancer Cytopathol. 2017;125(9):701‐709. [DOI] [PubMed] [Google Scholar]
- 30. Mukherjee M, Radio S, Wedel W, et al. Investigation of scanning parameters for thyroid fine needle aspiration cytology specimens: a pilot study. J Pathol Inform. 2015;6(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arnold MA, Chenever E, Baker PB, et al. The College of American Pathologists guidelines for whole slide imaging validation are feasible for pediatric pathology: a pediatric pathology practice experience. Pediatr Dev Pathol. 2015;18(2):109‐116. [DOI] [PubMed] [Google Scholar]
- 32. Donnelly A, Mukherjee M, Lyden E, et al. Optimal z‐axis scanning parameters for gynecologic cytology specimens. J Pathol Inform. 2013;4(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evered A, Dudding N. Accuracy and perceptions of virtual microscopy compared with glass slide microscopy in cervical cytology. Cytopathology. 2011;22(2):82‐87. [DOI] [PubMed] [Google Scholar]
- 34. Park S, Pantanowitz L, Parwani AV. Digital imaging in pathology. Clin Lab Med. 2012;32(4):557‐584. [DOI] [PubMed] [Google Scholar]
- 35. Girolami I, Pantanowitz L, Marletta S, et al. Diagnostic concordance between whole slide imaging and conventional light microscopy in cytopathology: a systematic review. Cancer Cytopathol. 2020;128(1):17‐28. [DOI] [PubMed] [Google Scholar]
- 36. Pantanowitz L, Wiley CA, Demetris A, et al. Experience with multimodality telepathology at the University of Pittsburgh Medical Center. J Pathol Inform. 2012;3(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: a systematic review. Arch Pathol Lab Med. 2017;141(1):151‐161. [DOI] [PubMed] [Google Scholar]
- 38. Araújo ALD, Arboleda LPA, Palmier NR, et al. The performance of digital microscopy for primary diagnosis in human pathology: a systematic review. Virchows Arch. 2019;474(3):269‐287. [DOI] [PubMed] [Google Scholar]
- 39. Khalbuss WE, Pantanowitz L, Parwani AV. Digital Imaging in Cytopathology. Patholog Res Int. 2011;2011:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Girolami I, Marletta S, Pantanowitz L, et al. Impact of image analysis and artificial intelligence in thyroid pathology, with particular reference to cytological aspects. Cytopathology. 2020;31(5):432‐444. [DOI] [PubMed] [Google Scholar]
- 41. Marletta S, Treanor D, Eccher A, Pantanowitz L. Whole‐slide imaging in cytopathology: state of the art and future directions. Diagn Histopathol. 2021;27(11):425‐430. [Google Scholar]
- 42. Cross S, Furness P, Igali L, Snead D, Treanor D. Best Practice Recommendations for Implementing Digital Pathology. Royal College of Pathologists; 2018. https://www.rcpath.org/uploads/assets/f465d1b3‐797b‐4297‐b7fedc00b4d77e51/Best‐practice‐recommendations‐for‐implementing‐digital‐pathology.pdf. Accessed June 21, 2018. [Google Scholar]
- 43. Campbell WS, Talmon GA, Foster KW, Baker JJ, Smith LM, Hinrichs SH. Visual memory effects on intraoperator study design: determining a minimum time gap between case reviews to reduce recall bias. Am J Clin Pathol. 2015;143(3):412‐418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


