Fig. 3.
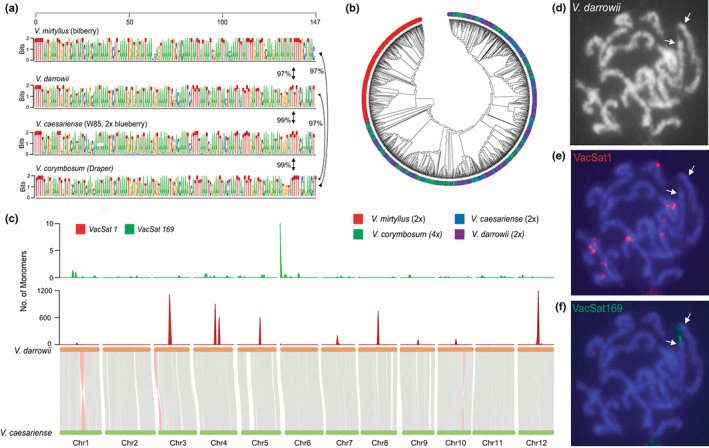
Comparative analysis of putative satellite centromeric repeat VacSat1 (147 bp monomer) in Vaccinium species. (a) VacSat1 sequence logo representing monomers extracted from four Vaccinium species, Draper (tetraploid, V. corymbosum), W85 (diploid, V. caesariense), bilberry (diploid, V. myrtillus) and evergreen blueberry (diploid, V. darrowii). Numbers next to the arrows indicate the average percent similarity of VacSat1 monomers across species estimated using the Maximum Composite Likelihood. (b) Phylogenetic analysis of VacSat1 monomer sequences inferred using the Neighbor‐Joining method implemented in Mega11 (Tamura et al., 2021). For the sequence logo and phylogenetic analysis, 300 VacSat1 monomer sequences/species were used. (c) Synteny analysis between W85_v2 against V. darrowii genome and distribution of VacSat1 and VacSat169 in V. darrowii physical maps (1 Mb windows). (d–f) Localization of VacSat1 and VacSat169 repeats on V. darrowii chromosomes using fluorescence in situ hybridization (FISH). (d) A somatic metaphase chromosomes of V. darrowii hybridized with (e) VacSat1 (red signals) and (f) VacSat169 (green). Arrows (in d and e) indicate the chromosomes with a terminal VacSat169 signal and no detectable VacSat1 repeats.
