Fig. 6.
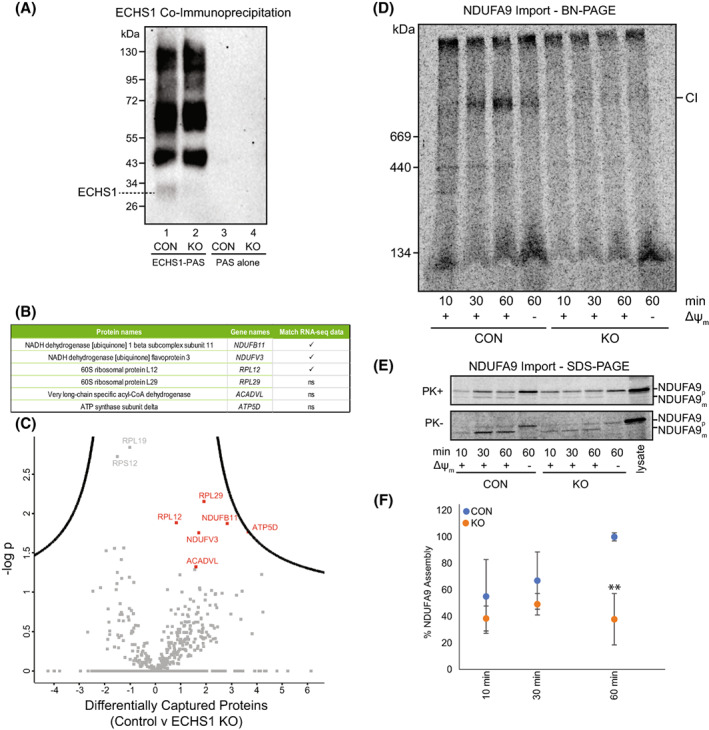
Co‐immunoprecipitation of ECHS1 identifies putative OXPHOS complex I binding partners, with loss of ECHS1 disrupting complex I assembly. Co‐immunoprecipitation with ECHS1 antibodies (ECHS1‐PAS) or PAS alone, followed by western blotting and immunodetection with anti‐ECHS1 antibodies. (A) ECHS1‐PAS pull‐down captured ECHS1 (Lane 1) from control (CON) mitochondria, with non‐specific proteins also pulled down (lane 1). ECHS1 was not captured from ECHS1 KO mitochondria as expected, however non‐specific proteins were pulled down (lane 2). PAS alone did not capture any non‐specific proteins detectable with anti‐ECHS1 antibodies (Lanes 3 and 4) from either CON or ECHS1 KO mitochondria. Image shown is representative of three independent experiments. (B) Six proteins specific for ECHS1 immunocapture identified by comparative analyses of co‐immunoprecipitated proteins captured from CON mitochondria with those captured from ECHS1 KO mitochondria. Of these, RNA‐seq identified reduced transcript levels of NDUFB11, NDUFV3 and RPL12 (ns, not significant). (C) Volcano plot of captured proteins by ECHS1 coimmunoprecipitation. Permutation‐based FDR set at < 1% and s0 = 1. RPL12, RPL29, NDUFB11, NDUFV3, ATP5D and ACADVL (shown in red) present in capture from CON mitochondria but not ECHS1 KO mitochondria (n = 3, Student's two‐tailed t‐test, P < 0.05). (D) BN‐PAGE showing the assembly of NDUFA9 into mature complex I (CI) following solubilisation of 40 μg mitochondria per lane in 1% TX‐100. (E) SDS/PAGE showing NDUFA9 following import into both CON and ECHS1 KO mitochondria. NDUFA9 is detectable in its precursor (p) form and as a proteinase K (PK+) resistant mature (m) form. (F) Quantitation of NDUFA9 assembly into mature complex I after 10, 30 and 60 min of import, normalised to maximum amount of NDUFA9 imported after 60 min in CON. The amount of NDUFA9 assembled into complex I (CI) after 60 min in ECHS1 KO mitochondria was only 37.9 ± 19.3% of CON levels. Data shown are mean ± SD with n = 3, **P < 0.01 relative to control (CON; Student's two‐tailed t‐test). Images shown are representative of three independent experiments.
