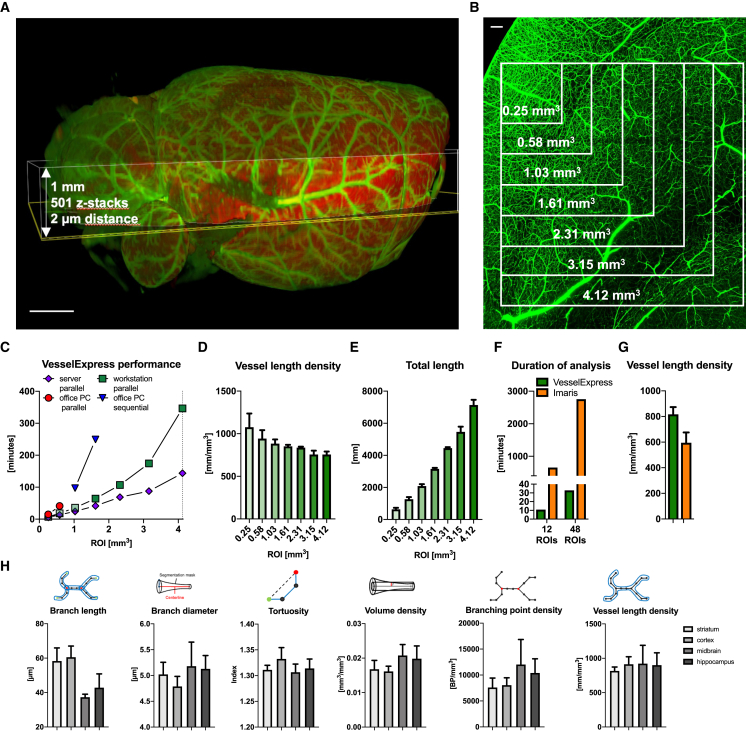
Figure 2.
Analysis of FITC-albumin labeled brain vessels using automated pipeline VesselExpress
(A) Overview of brain vasculature in a mouse brain scanned with LSFM. Highlighted area represents region in which detail scans for vessel analyses in (C–G) were performed.
(B) Maximum intensity projection of detailed image from LSFM scan using 6.64× magnification and a step size of 2 μm. Inlets showing the region of interest used for vascular analysis in (C–E).
(C) Duration of vascular analysis in VesselExpress using four ROIs each in parallel with increasing image sizes. Analysis was performed on different devices. Using an office PC containing an Intel Core Duo i9 processor with 2.40 GHz, 8 cores, and 32 GB RAM, parallel analysis was possible with ROIs of a maximum size of 0.58 mm3 (red circles). Using one ROI per run, it was possible to analyze a ROI with 2.31 mm3, and total run times were summed up (blue triangle).
(D and E) Green squares represent a workstation equipped with an Intel Xeon W-1255 CPU with 3.30 GHz and 512 GB RAM, whereas violet diamonds display runtime of the very same ROIs on a server working with an Intel Xeon E7-8890 v4 CPU with 2.20GHz, 96 cores, and 1.97 TB RAM. Results from this analysis are displayed as vessel length density (D) and total vessel length (E).
(F and G) Duration (F) and vessel length density (G) from the analysis of either 12 ROIs or 48 ROIs with an image size of 0.25 mm3 using the automated pipeline VesselExpress or sequentially analysis using Imaris software (Bitplane, Oxford Instruments).
(H) Analysis of FITC-albumin-labeled vascular structure performed in 4–12 ROIs of striatum, cortex, midbrain, hippocampus, and corpus callosum of brains from up to six male mice using VesselExpress. Data in (D), (E), (G), and (H) are means ± standard deviation values. Scale bars represent 1 mm in (A) and 100 μm in (B).