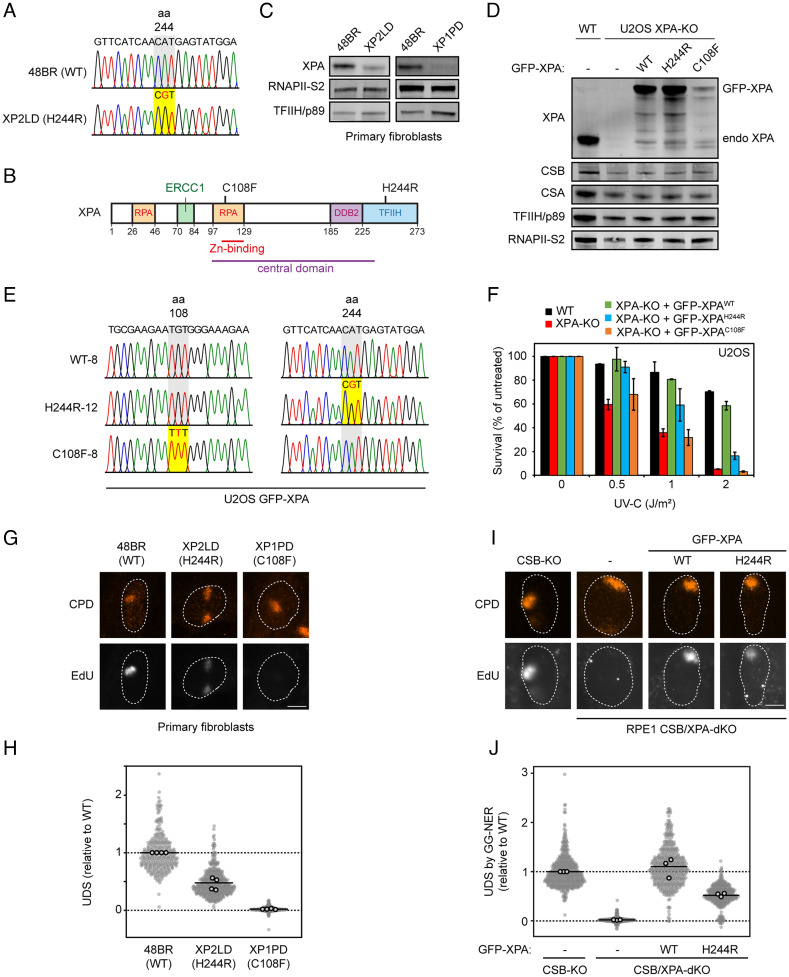
Fig. 1.
Patient or rescue cells expressing XPAH244R show residual UDS. (A) Sequence confirmation of the H244R amino acid substitution in genomic DNA of patient XP2LD compared to WT DNA (48BR). (B) Schematic representation of the XPA protein with its interaction domains. (C) XPA protein expression in primary fibroblasts from 48BR (WT), XP2LD (XPAH244R), and XP1PD (XPAC108F) by western blot. RNAPII-S2 and TFIIH/p89 levels are used as loading controls. (D) Expression levels of endogenous XPA in U2OS(FRT) WT and U2OS(FRT) XPA-KO cells, and GFP-XPA expression in U2OS(FRT) XPA-KO cells reconstituted with GFP-XPAWT, GFP-XPAH244R, or GFP-XPAC108F. CSB, CSA, TFIIH/p89, and RNAPII-S2 are used as loading controls. (E) Sequence confirmation of the H244R or C108F amino acid substitution in the U2OS(FRT) XPA-KO cells reconstituted with GFP-XPAWT, GFP-XPAH244R, or GFP-XPAC108F cDNA. (F) Clonogenic survival assay after irradiation with various doses of UV-C in U2OS(FRT) WT, XPA-KO, and GFP-XPA reconstituted cell lines. The bars are the average of two experiments. The error bars reflect the SD. (G) Representative images and (H) quantification of EdU incorporation in the indicated primary fibroblasts after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by CPD staining. The EdU signal has been normalized for CPD levels at sites of local damage. (I) Representative images and (J) quantification of EdU incorporation in the indicated RPE1 cells deficient in TC-NER (CSB-KO cells) to specifically measure UDS by GG-NER after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by CPD staining. The EdU signal has been normalized for CPD levels at sites of local damage. (H and J) All cells are depicted as individual data points with the bar representing the mean of all data points. The individual means of four biological replicates are depicted as gray points with black circles.