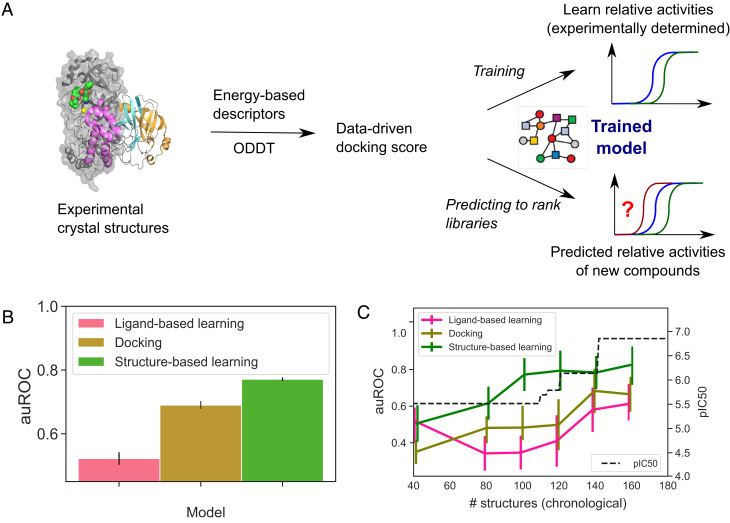
Fig. 2.
Structure-based learning outperforms docking and ligand-based machine learning in relative affinity predictions. (A) To build a model that captures the relationship between the crystal structure of a protein–ligand complex and the activity of the ligand, we extracted Autodock Vina descriptors from each crystal structure and deployed it to predict the relative potency of ligands. (B) Comparison of this approach (green) with ligand-based learning (pink) and docking (yellow) using data from the COVID Moonshot campaign and a classification task that evaluates pairwise comparisons between the activities of ligands using a cutoff of 0.5 log10 units for classifying one compound as more active than another. The average auROC scores were computed using a scaffold split, with the error bars corresponding to SE of the mean. Performance on individual scaffolds is shown in Dataset 4. (C) Time-split evaluation of our model. Our approach maintains its predictive power when trained on older, less potent molecules and asked to rank the affinities of newer more potent molecules (green line). Error bars correspond to SE of the mean. The black line tracks the potency of the most potent molecule discovered at that point.