Figure 10.
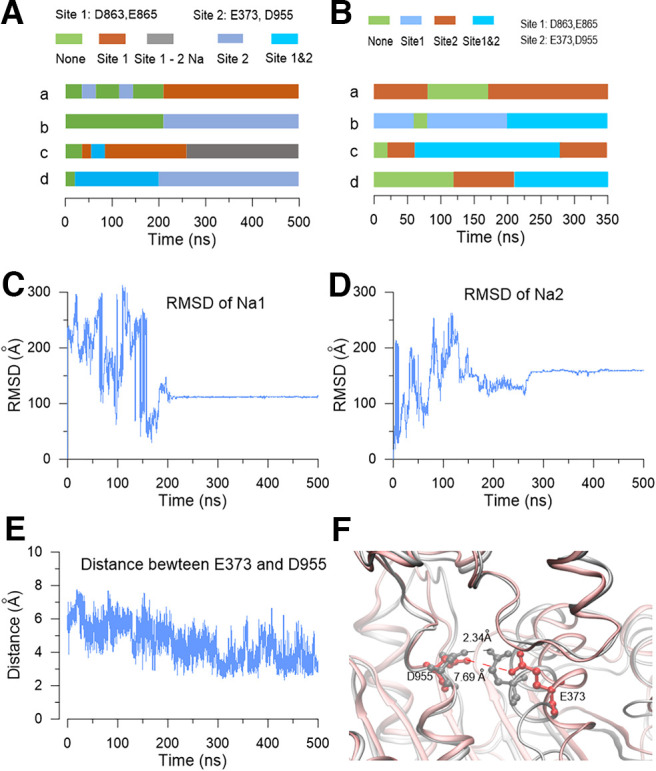
The time of the resident sodium ions staying in the sodium-sensitive site of the rSlack channel during the MD simulation. A, The time of four subunits with one or two sodium sites occupied or vacant in the homology model during the 500 ns MD simulation was indicated by different colors. B, The time of the four subunits with one or two sodium sites bound by one or two sodium ions and one vacant site during the 350 ns MD simulation. C, D, The rmsD of the two sodium ions (Na1 and Na2) binding to the E373/D955 site during the 500 ns MD simulation. E, The distance between the E373 and D955 site of one subunit during the 500 ns MD simulation. F, The snapshots of the local structure of the E373/D955 site with (gray) or without (red) sodium binding.
