Figure 16.
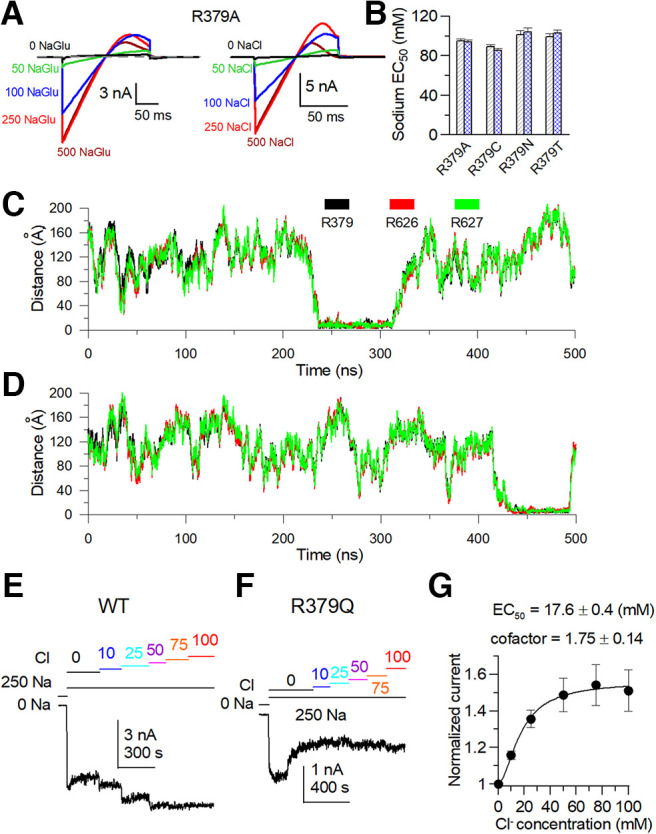
Distances of Cl– to positively charged amino acids R379Q and R626/627Q of rSlack channel in the simulated local structural model. A, Sample traces of R379A in NaGlu and NaCl solution. B, Comparison of sodium sensitivity of the R379 mutant EC50 values in NaGlu and NaCl solution. The sodium dependence of the R379 mutant EC50 value in NaGlu are as follows (in mm): R379A, EC50 = 95 ± 2; R379N, EC50 = 89 ± 2.1; R379C, EC50 = 101.3 ± 4.2; and R379T, EC50 = 99.3 ± 3.3. The EC50 values in NaCl are as follows (in mm): R379A, EC50 = 94 ± 1.6; R379N, EC50 = 85.4 ± 4.4; R379C, EC50 = 104 ± 7.3; and R379T, EC50 = 102.8 ± 4.2. C, During the simulation process, distances of the bound Cl– ion to residues R379, R626, and R627 amino acids. D, Distances of another bound Cl– ion to residues R379, R626, and R627 amino acids during the simulation process. E, F, The time course of the current change of the WT Slack channel and R379Q mutant were perfused in different Cl– and 250 mm sodium solution concentrations. G, The Cl– dose-dependent curve for the activation of the Slack channel.
