Figure 7. Potent HCV-neutralizing activity requires a pattern of genetic features and can be predicted.
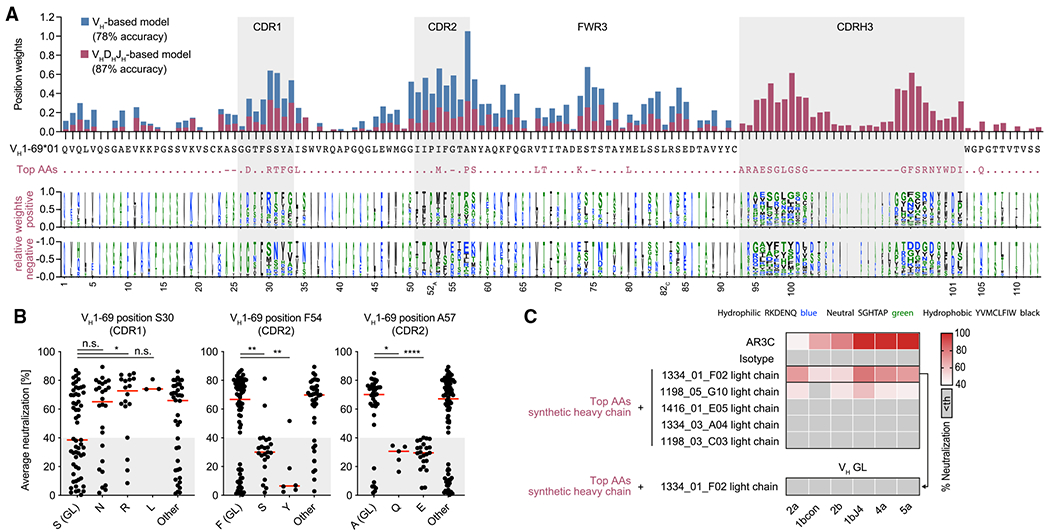
(A) Top panel: total weights of VH1-69-amino-acid positions contributing to the classification as an efficient neutralizer in VH-based or VHDHJH-based machine learning models. The VH1-69*01 germline amino acids are shown in black and the most impactful amino acids for the VHDHJH-based model (top AAs) are shown in violet. Bottom panel: logo plot with letter heights representing relative weights of amino acids at each position for positive or negative classification of the encoded antibodies as efficient neutralizers. Letter widths represent absolute weights. Insertions are omitted for clarity.
(B) Average neutralization values of antibodies with indicated amino acid sequence features in the HCVcc screen shown in Figure 3A. Lines indicate medians.
(C) HCVcc neutralization by antibodies with a synthetic heavy-chain amino acid sequence as depicted in (A) in violet. Medians of triplicate measurements. Representative of 2 independent experiments. Th, threshold.
Not significant (n.s.), p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001; two-tailed Mann-Whitney test with Bonferroni correction. GL, germline.
