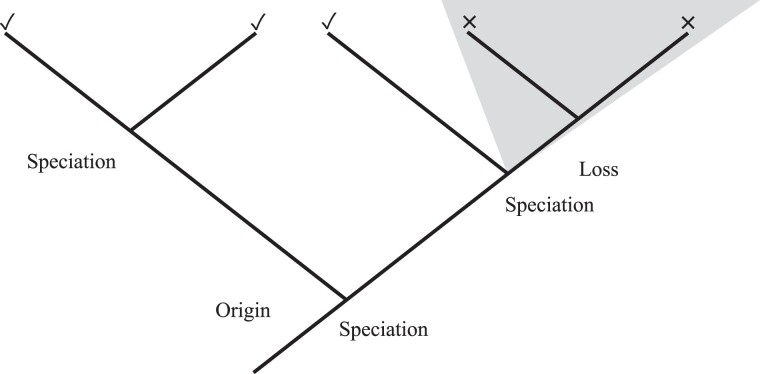
Fig. 5.
Inferring loss events for a Pfam by Dollo parsimony. Dollo parsimony assumes a Pfam arose exactly once, on the branch basal to the monophyletic clade that includes all species with the Pfam, and is lost on the branches that explain the current distribution with the fewest number of losses. Species with at least one instance of the Pfam are indicated with checks and those without by crosses. We initialize the Pfam loss rate as 1/sum of unshaded branch lengths, that is, all unshaded branches are counted by index k, whereas the most basal-shaded branch is counted by index j in equation (1). We then refine this coarse Dollo parsimony-based estimate through ML techniques that allow for false positive/horizontally transferred instances (eq. 2).